• Discovered ~20 years ago, aptamer applications are booming
• Modified oligos and nucleotide triphosphates play pivotal roles
• World’s first chemical guided missile could be the answer to wiping out cancer
• Are antibodies passé?
Aptamers are nucleic acids (or peptides) that bind to a specific target molecule. RNA or DNA aptamers are usually created by selection from a large pool (aka library) of random sequences; however, natural RNA aptamers also exist in riboswitches. As discussed below, aptamers have been used for an impressively wide variety of applications in either basic research or, especially, health-related diagnostics and therapeutics. This remarkable utility is clearly reflected in the publication statistics—since their discovery in 1990, there have been ~11,000 publications indexed to DNA or RNA aptamers in SciFinder with a projected average rate of ~5 per day in 2013! More than 1,600 of these publications are patents, which is a stunning testament to the commercial potential of aptamers.
Most published authors: Analyzing these SciFinder aptamer publications by author gave the following top-5 ranking for number of publications indexed to aptamers…amazing productivity!
Tan, Weihong: 306
Ellington, Andrew D.: 173
Ikebukuro, Kazunori: 112
Famulok, Michael: 99
Sullenger, Bruce A.: 97
Trying to get what you want sometimes leads to getting what you need…
One concept, two labs, and a pair of seminal publications
By remarkable coincidence in 1990, two labs independently published conceptually similar achievement of RNA selection in vitro—without using an RNA replicase, as in the past—but by quite different approaches.
Craig Tuerk and Larry Gold described in Science a procedure they cleverly named SELEX, which sounds a bit like select, and was an acronym for systematic evolution of ligands by exponential enrichment. This now classic paper has nearly 5,000 citations in Google Scholar. The authors noted that “[t>
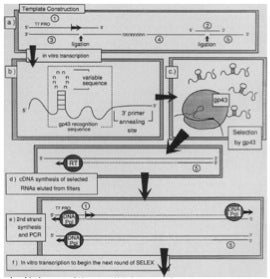
he method relies on mechanisms usually ascribed to the process of evolution, that is, variation, selection, and replication.” Briefly, as depicted in the figure below taken from this paper, a segment of RNA was transcribed from DNA template that had been synthesized so as to have eight contiguous, “randomized” positions (N = A, G, C and T) by means of “mixed couplings” using all four phosphoramidite reagents. This pool of 65,536 ( = 48) different RNAs was incubated with gp43 and the subset of RNAs selected by gp43 were then collected, reverse-transcribed into cDNA, amplified by PCR, and then transcribed into RNA for a second round of SELEX. After six rounds of SELEX, sequencing 20 clones of cDNA revealed that, in addition to the wild type 8-nt RNA sequence, a major variant mutated at 4-of-8 positions was also selected, along with several minor variants with fewer mutations.
Those interested in the scientific context of the above work, and the molecular biological interpretations, are encouraged to read the original paper, which is freely available as a pdf through a Google Scholar by searching for “Tuerk Gold 1990”. Here, however, are selected—pun intended—prescient comments by Tuerk and Gold in their concluding section simply entitled “Applications.”
• “Among many general applications of this technique, SELEX analyses can be used to determine the optimal binding sequences for any nucleic acid binding protein.”
• “Small molecules…can be bound to insoluble supports to partition RNAs that interact specifically with these substrates.”
• “We expect that, at the very least, nucleic acid ligands that inhibit replicative proteins of epidemiologically important infections can be likewise evolved. Other, more sophisticated effects of evolved RNA molecules on target activities may be possible."
• “Finally, SELEX may be just the beginning of evolution in a test tube…[and>
provide unpredictable and unimaginable molecular configurations of nucleic acids and proteins with any number of targeted functions.”
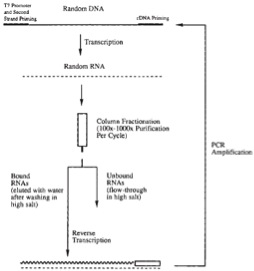
Andrew D. Ellington and Jack W. Szostak reported in Nature Methods the synthesis of large numbers of random sequence RNA molecules for binding to specific ligands to investigate fundamental questions related to formation of stable polynucleotide 3-dimensional structures in the context of theories of the origin and early evolution of life. Briefly, they synthesized ~150-nt DNAs comprised of central ~100-nt random sequence—via mixed phosphoramidite couplings (see above)—flanked by defined-sequence 5’ and 3’ ends as primer and restriction sites for cloning. As depicted below in a figure from this paper, transcribed random-sequences of RNA were applied to a column of agarose having an immobilized dye as a model ligand.
They chose six dyes that have “many possible hydrogen-bond donor and acceptor groups as well as planar surfaces for stacking interactions.” Bound RNAs were eluted and reverse transcribed for PCR amplification before another cycle of selection. After four to five cycles and sequencing, it was concluded that binding of selected RNAs is specific with regard to cross-reactivity of each selected pool for each column. However, each dye-binding pool was estimated to be a complex mixture of 102 – 105 different sequences, i.e. “there are many independent sequence ‘solutions’ to each ligand-binding ‘problem.’“
For some reason, reading this analogy of many solutions for one problem brought to mind these lyrics from the song in the Rolling Stones 1969 album Let It Bleed:
No, you can't always get what you want
But if you try sometime, you just might find
You get what you need
Those interested in the details of clonal sequence information, etc. extracted from the results are encouraged to read the original paper, which is freely available as a pdf through a Google Scholar search for “Ellington Szostak 1990.” These authors went on to say that “[w>
e have termed these individual RNA sequences ‘aptamers’, from the Latin ‘aptus’, to fit. Furthermore, they suggested that “it may be possible to isolate novel ribozymes from pools of random-sequence RNAs” using transition state analog affinity columns, by analogy with the isolation of catalytic antibodies.
The enabling power of aptamers
The vast body of literature covering basic research and numerous applications of nucleic acid aptamers is a stunning testament to the enabling power of these molecules, and the myriad of expanded types of aptamers and aptamer-generation methods. The Ellington Lab has provided a very useful tool for obtaining information about apatamers in the form of The Aptamer Database, which is a comprehensive, annotated repository for information about aptamers and in vitro selection. This searchable resource is provided to collect, organize and distribute all the known information regarding aptamer selection. For example, when I searched for “toxin” in Title Keywords, three citations were provided with active links, e.g. this 2006 article by Tang et al. in Electrophoresis: The DNA aptamers that specifically recognize ricin toxin are selected by two in vitro selection methods.
Additional search tools for aptamer-related publications, such as PubMed, Scirus, and SciFinder, readily provide access to whatever aspects may be of interest. In perusing this wealth of information, I came across the following items that I selected—pun intended (again!)—as representative examples of recently available literature for various categories.
Selections Methods:
• Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer
• Multiplexed microcolumn-based process for efficient selection of RNA aptamers
• In vivo SELEX for Identification of Brain-penetrating Aptamers
Targeting Cells:
• Development of an Efficient Targeted Cell-SELEX Procedure for DNA Aptamer Reagents
• Selection of DNA Aptamers against Glioblastoma Cells with High Affinity and Specificity
Therapeutics:
• Nucleic acid aptamers: clinical applications and promising new horizon
• Therapeutic RNA aptamers in clinical trials
• An RNA Alternative to Human Transferrin: A New Tool for Targeting Human Cells
Diagnostics:
• Development of an Aptamer-Based Concentration Method for the Detection of Trypanosoma cruzi in Blood
• Harnessing aptamers for electrochemical detection of endotoxin
Food Safety:
• Aptamer-Based Molecular Recognition of Lysergamine, Metergoline and Small Ergot Alkaloids
• Nucleic acid aptamers for capture and detection of Listeria spp
“World’s first chemical guided missile could be the answer to wiping out cancer”
This attention grabbing headline of an article in Deakin University Newsroom led me to contact Prof. Wei Duan regarding the current status of this promising research that he and his team first described in a Cancer Sci. 2011 publication entitled RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. In this study, SELEX was used to identify a 40-base RNA aptamer that binds to epithelial cell adhesion molecule (EpCAM), which is overexpressed in most solid cancers and is reported to be a cancer stem cell marker. Interestingly, the aptamer was further truncated to 19 bases yet still interacts specifically with a number of live human cancer cells derived from breast, colorectal, and gastric cancers that express EpCAM, but not with those not expressing EpCAM. Importantly, this EpCAM RNA aptamer is efficiently internalized after binding to cell surface EpCAM. These authors believed that this is the first RNA aptamer against a cancer stem cell surface marker being developed, and that such cancer stem cell aptamers will greatly facilitate the development of novel targeted nanomedicine and molecular imaging agents for cancer theranostics.
Prof. Wei Duan, Director of Nanomedicine, Deakin University (Geelong, Victoria, Australia); taken from Deakin University website via Bing Images.
As for the current status of this line of research, Prof. Duan provided me with the following comments. “With a few rare exceptions, most of the chemotherapy drugs and molecularly targeted agents in oncology clinics kill only non-cancer stem cells. In order to improve the cancer treatment outcome, we must target cancer stem cells. Our lab has developed the world’s first RNA aptamer against cancer stem cell marker protein. We are currently working on novel treatment strategies by targeting cancer stem cells in vivo using our aptamers. For example, using cancer stem cell targeting aptamers, we can transform conventional chemotherapy drugs to robust cancer stem cell busters in xenograft tumour models. With some smart engineering and strategic chemical modifications, we can now deliver siRNA into cancer stem cells in vivo to achieve an unprecedented %ID (percentage of injected dose). I believe that furture development of aptamer for cancer stem cell targeting will transform the way we think and approach cancer treatment.”
Expanding chemical modifications to an expanded genetic alphabet
Moving beyond sequence pools comprised of natural A, G, C, and T/U to chemically modified nucleotides in aptamers has been extensively investigated to either increase diversity of structural elements (e.g. hydrophobicity, polarity, 3D shape, etc.) to obtain improved binding, or improve biological properties (e.g. nuclease resistance, cellular uptake, etc.). Because of limitations in both enzyme activity and the modified NTPs readily available, the vast majority of modified aptamers have been developed using 2′ fluoro dC (fC) and 2′ fluoro dU (fU), although the 2′ amino and 2′ OMe analogs of the pyrimidines have also been used. More “exotic” modified dNTPs with pendant amino acid or hydrophobic moieties that have been investigated can be perused by clicking here for a 2012 review by Marcel Hollenstein entitled Nucleoside Triphosphates — Building Blocks for the Modification of Nucleic Acids.
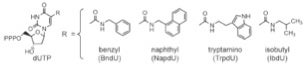
Especially noteworthy, however, is SomaLogic’s development of novel SOMAmer™ (Slow Off-rate Modified Aptamer) protein-binding technology capable of measuring thousands of proteins in small volumes of biological samples with low limits of detection, a broad dynamic range, and high reproducibility. This approach, which was first published in 2010 in PLoS One, uses chemically-modified nucleotides shown below to mimic amino acid side chains together with new SELEX strategies. SOMAmers have already been developed for more than 1,100 different protein targets critical to normal and disease biology, with many more said to come soon.
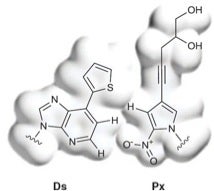
More recently, a truly remarkable milestone in aptamer technology was reported earlier this year in Nature Biotechnology by Ichiro Hirao and coworkers at the RIKEN Systems and Structural Biology Center and TagCyx Biotechnologies in Japan. These investigators note that DNA aptamers produced with natural or modified natural nucleotides often lack the desired binding affinity and specificity to target proteins. They then describe a method for selecting DNA aptamers containing the four natural nucleotides and an unnatural nucleotide with the hydrophobic base Ds, which specifically pairs with the unnatural base Px during PCR, as depicted below and highlighted in an earlier related blog.
Up to three Ds nucleotides were incorporated in a random sequence library. Selection experiments against two human target proteins, vascular endothelial cell growth factor-165 (VEGF-165) and interferon-γ (IFN-γ), yielded DNA aptamers that bind with KD values of 0.65 pM and 0.038 nM, respectively, affinities that are >100-fold improved over those of aptamers containing only natural bases. The authors concluded that “[t>
hese results show that incorporation of unnatural bases can yield aptamers with greatly augmented affinities, suggesting the potential of genetic alphabet expansion as a powerful tool for creating highly functional nucleic acids.”
Verified Randomized Sequences
I highly recommend reading Ellington and coworkers Current Protocols in Nucleic Acid Chemistry (2009) for detailed guidance on design, synthesis, and amplification of DNA pools for in vitro selection. These experts note that the “quality of randomization” is critically dependent on various aspects of automated DNA chain assembly using empirically determined ratios of A, G, C and T phosphoramidites to generate each N. Completely random pools should theoretically have 25% base composition at each N position, and therefore an unbiased distribution of di-, tri-, etc. nucleotide sequences.
If you’re looking for a supplier for verified random sequences, check out TriLink Biotechnologies. TriLink is a leader in verified manufacturing of randomized sequences that are now offered as convenient, stocked DNA or RNA random-libraries having 20, 30 or 40 randomized Ns and restriction sites for use with corresponding PCR primers and biotin labeling if desired. The mix of individual bases has been optimized to ensure as close to a 1:1:1:1 ratio of bases (i.e. 25% base composition) as possible during oligo synthesis. Each lot is tested by enzymatic digest to determine overall base composition and to verify that it falls within TriLink's rigorous specifications, ensuring lot-to-lot reproducibility.
To my knowledge, TriLink is the only supplier of randomized sequences that have been verified by high-throughput applications (aka next-generation sequencing) to more deeply analyze randomization. NeoVentures BioTechnology has independently sequenced 61 clones of a 40-N library to obtain base composition at each position and, importantly, tri-nucleotide distribution, as detailed in this white paper. By contrast, NeoVenture’s analysis of a sample from an anonymous vendor revealed substantial deviation from random base composition and, consequently, unacceptable skewing of tri-nucleotide distribution.
Does complete randomness matter? Experts say yes: Ellington and coworkers opine that “[c>
ompletely random sequence pools are used to initiate selection experiments when no functional nucleic acid sequence or structural motif is known in advance.” Furthermore, they add, “completely random sequence pools explore a much wider swath of sequence space and are more useful for the isolation of novel binding species (aptamers) or catalytic species.”
Commercial Landscape
In July 2013 a comprehensive and up-to-date report entitled Aptamers Market - Technology Trend Analysis By Applications - Therapeutics, Diagnostics, Biosensors, Drug Discovery, Biomarker Discovery, Research Applications with Market Landscape Analysis - Global Forecasts to 2018 was made available by Market Research.com for purchase. The following is included in the free abstract of this report.
• Aptamers is an emerging market, widely considered as a rival or substitute to antibodies in the scientific industry. It is poised to grow rapidly in various application areas, including therapeutics and diagnostics. The global aptamers market is valued to be $287 million in 2013 and is expected to reach $2.1 billion by 2018.
• The list of promising aptamers in the clinical trials pipeline estimates that these synthetic chemical antibodies will soon surpass monoclonal antibodies in therapeutics, diagnosis, and imaging. With technological merits over antibodies, the aptamers market is poised to grow at par with antibodies in the next 10-15 years.
• The advantages of chemically modified aptamers and low production cost compared to antibodies aid the growth of the market.
• There are ~60 biotech and pharmaceutical companies working on aptamers.
A review of nanotechnology and aptamers noted an important linkage between “expiration of the early patents…and a simultaneous decentralization of aptamer-technology commercialization”.
Given the range of applications, growth trajectory, and estimated market potential of aptamers, I’m guessing this is a field many of you currently are or will want to monitor. I’ve put together a list of notable players to keep your eye on. These include, in alphabetical order: AM Biotechnologies, LLC (U.S.) Aptagen, LLC (U.S.) Aptamer Sciences, Inc. (S. Korea) Aptamer Solutions, Ltd. (U.K.) Base Pair Biotechnologies (U.S.), Bioapter S.L. (Spain), Integrated Biotechnologies, Inc. (U.S.), NeoVentures Biotechnology, Inc. (Canada), NOXXON Pharma AG (Germany), OTC Biotech (U.S.), Regado Biosciences, Inc. (U.S.), and TriLink BioTechnologies (U.S.)—hum, maybe I should have done this list in reverse alphabetical order.
What lies ahead?
I don’t believe in a crystal ball that can predict the future for aptamers, or anything else for that matter. After searching the literature for aptamers and keywords such “future”, “perspective”, “horizon” or other forward-looking terms, I decided that there wasn’t any publication that I could quote from and cite as providing insights as to new directions. Consequently, I’ll offer my half-thoughts on some things that might occur at some time in the future.
Overall, I think that applications of aptamers will out-pace antibodies based on inherently greater structural diversity that can be obtained chemically, as opposed to biologically, and with a much more limited set of amino acid/analog “building blocks”. Other advantages, as noted by Base Pair Biotechnologies, include more economical production—especially in GMP grade—flexible “fine tuning” for desired bio-distribution, stability or shelf-life.
High-throughput sequencing and related bioinformatics will likely continue to enable new and improved aptamer selection methods, as exemplified by the recent work of Hoon et al., who used one such approach to develop a novel method for selecting high-affinity DNA aptamers based on a single round of selection.
Aptamers—not antibodies—will advantageously be selected for small molecule targets to develop new or improved label-free electronic sensors, such as those reported by Ohno et al. for an aptamer-modified graphene field-effect transistor. Incidentally, ethanolamine (HOCH2CH2NH2), which is associated with several diseases and has only four non-hydrogen atoms, was reported by Mann et al. to be perhaps the smallest aptamer target so far.
Finally, I expect that molecular modeling will get faster, cheaper, better and more accessible to folks wanting an aptamer against a molecular target, and thereby enable more computationally guided aptamer selection. One example of advances in this direction can be found in a 2013 publication entitled Computational approaches to predicting the impact of novel bases on RNA structure and stability by Harrison et al.
What do you think? Your ideas or other comments on this post are welcomed.
Postscript: Latest breaking aptamer news
News from Naples
This first ever international meeting on Aptamers In Medicine Perspectives, which was supported in part by TriLink, brought together many experts in, and practitioners of aptamer technology whose presentations are listed in the Final Program. TriLink's CEO, Rick Hogrefe was in attendance and provided the following insights:
“Dr. Hoinka, of the National Center for Biotechnology Information, described the use of a novel high- throughput (HT) SELEX method called HTSAptamotif, which is coupled to an NGS platform and probably a much bigger computer than most labs have, to crunch out sequence and motif patterns that allow you to rationally design your aptamer. It is reasonable to expect that some year all that will be needed to design the perfect aptamer will be the crystal structure of the target and your somewhat smarter smart phone. An iPhone app will be soon to follow, no doubt.”
Rick also observed that “[o>
ne reoccurring topic of discussion was the advantages of using chemically synthesized aptamers versus those made by transcription. One of the main disadvantages of using long RNA aptamers (>60mer) is that they must be prepared by transcription. Methods are still in development to prepare these compounds economically and eventually at scale. Many researchers truncate their original sequence to something more manageable—a 19-mer was reported in one talk. The greatest fear regarding the use of enzymatic synthesis of aptamers was the removal of one very deleterious contamination—5’-triphosphate oligonucleotides, which are potent activators of RIG-I. This is done with a phosphatase after transcription. During the Q&A session that followed the talk of Dr. Zhou of John Rossi’s lab at the City of Hope, Dr. Sullenger said he very carefully studied this reaction and was unable to optimize it sufficiently to remove all the triphosphates from the aptamer. Another attendee believed that they were able to sufficiently remove the triphosphate. More work is needed to develop a good method to detect the presence of triphosphate modifications on long oligos.”
Rick’s concluding opinions are that “[o>
verall, it is apparent that aptamers are not only here to stay, but are rapidly expanding into what may become the most universally applicable oligonucleotide technology to date. The excellent talks in this meeting covered much of the breadth of what aptamers are capable of doing. It is time to buy your random library kit from TriLink and start your own aptamer research. Most likely you will find an aptamer that fits the bill—and your target—perfectly.”
Where are they now?
 Dr. Gold founded SELEX-based NeXagen, which later became NeXstar Pharmaceuticals. Gilead acquired NeXstar in 1999 and teamed up with (OSI) EyeTech to develop Macugen (aka Pegaptanib), which is an anti-angiogenic, chemically modified RNA aptamer for the treatment of neovascular (wet) age-related macular degeneration. Gold is the founder, chairman of the board, and CEO of SomaLogic, where he continues his work on aptamers. Dr. Gold was made a member of the American Academy of Arts and Sciences in 1993, and the National Academy of Sciences in 1995.
Dr. Gold founded SELEX-based NeXagen, which later became NeXstar Pharmaceuticals. Gilead acquired NeXstar in 1999 and teamed up with (OSI) EyeTech to develop Macugen (aka Pegaptanib), which is an anti-angiogenic, chemically modified RNA aptamer for the treatment of neovascular (wet) age-related macular degeneration. Gold is the founder, chairman of the board, and CEO of SomaLogic, where he continues his work on aptamers. Dr. Gold was made a member of the American Academy of Arts and Sciences in 1993, and the National Academy of Sciences in 1995.
 Craig Tuerk was a graduate student with Dr. Gold, and then worked at NeXagen (a SELEX-based company founded by Gold). Dr. Tuerk left NeXagen in 1994, many years before his co-invention of SELEX would ultimately lead to Macugen (see Gold above). He currently teaches biochemistry and genetics at Kentucky’s Morehead State University. The Tuerk Lab research interests include in vitro evolution strategies, combinatorial libraries and drug design.
Craig Tuerk was a graduate student with Dr. Gold, and then worked at NeXagen (a SELEX-based company founded by Gold). Dr. Tuerk left NeXagen in 1994, many years before his co-invention of SELEX would ultimately lead to Macugen (see Gold above). He currently teaches biochemistry and genetics at Kentucky’s Morehead State University. The Tuerk Lab research interests include in vitro evolution strategies, combinatorial libraries and drug design.
 The Szostak Lab is affiliated with the Center for Computational & Integrative Biology, Massachusetts General Hospital, the Department of Molecular Biology, Massachusetts General Hospital, the Department of Chemistry and Chemical Biology, Harvard University, the Department of Genetics, Harvard Medical School, and the Howard Hughes Medical Institute. The Nobel Prize in Physiology or Medicine 2009 was awarded jointly to Elizabeth H. Blackburn, Carol W. Greider and Jack W. Szostak "for the discovery of how chromosomes are protected by telomeres and the enzyme telomerase."
The Szostak Lab is affiliated with the Center for Computational & Integrative Biology, Massachusetts General Hospital, the Department of Molecular Biology, Massachusetts General Hospital, the Department of Chemistry and Chemical Biology, Harvard University, the Department of Genetics, Harvard Medical School, and the Howard Hughes Medical Institute. The Nobel Prize in Physiology or Medicine 2009 was awarded jointly to Elizabeth H. Blackburn, Carol W. Greider and Jack W. Szostak "for the discovery of how chromosomes are protected by telomeres and the enzyme telomerase."
 Dr. Ellington received his graduate degree in Biochemistry and Molecular Biology from Harvard University (working with Dr. Steve Benner), and was a post-doctoral Fellow with Jack Szostak at Massachusetts General Hospital. He was an assistant faculty member at Indiana University in the Department of Chemistry until 1998, and has since been a full Professor in the Department of Chemistry and Biochemistry at the University of Texas at Austin. Throughout his career Dr. Ellington has worked on the directed evolution of molecules and organisms, and has developed both insights into the origin of life and applications in biotechnology. The Ellington Lab motto is "what evolves here changes the world."
Dr. Ellington received his graduate degree in Biochemistry and Molecular Biology from Harvard University (working with Dr. Steve Benner), and was a post-doctoral Fellow with Jack Szostak at Massachusetts General Hospital. He was an assistant faculty member at Indiana University in the Department of Chemistry until 1998, and has since been a full Professor in the Department of Chemistry and Biochemistry at the University of Texas at Austin. Throughout his career Dr. Ellington has worked on the directed evolution of molecules and organisms, and has developed both insights into the origin of life and applications in biotechnology. The Ellington Lab motto is "what evolves here changes the world."
TagCyx Biotechnologies
According to its website, TagCyx Biotechnologies “core technology, the ‘unnatural base pair system’, has been developed by Dr. Hirao, founder and CEO of TagCyx Biotechnologies, during the course of his research at ERATO project (Exploratory Research for Advanced Technology) of Japan Science and Technology Agency, followed by the research at the University of Tokyo, and presently at RIKEN. The ‘Unnatural base pair system’ is an innovative technology which enables the site-specific incorporation of functional component into DNA, RNA and proteins. TagCyx Biotechnologies was founded in March 2007 as a RIKEN authorized venture business, with the mission of dissemination and application of this technology to various fields.”
From the clever corporate name joining natural base pairs TA and GC with variable, unnatural base pair YX, we can expect to see more from this company in the future.
Incidentally, according to a press release earlier this year, Dai Nippon Printing Co., Ltd. and TagCyx Biotechnologies have jointly developed a printing ink with a highly effective anti-counterfeit feature, through the integration of artificial DNA that is extremely difficult to replicate. This artificial DNA based ink is virtually impossible for third parties to replicate, and as it makes it possible to achieve powerful judgments of authenticity, can be used in anti-counterfeiting efforts connected with high-value printing requiring strong security, such as bank notes or cash vouchers.
MRI by DNA aptamers in vivo
After writing this blog, Philip Liu and his coworkers at Massachusetts General Hospital published the first-ever demonstration of protein-guided magnetic resonance imaging (MRI) by DNA aptamers in vivo using, as a model system, transcription factor (TF) protein AP-1, a heteroduplex protein comprising members of Fos and Jun immediate early gene families that plays an essential role in modifying gene expression by signal transduction. This work was published in FASEB J. A preprint of this report—kindly provided to me by Dr. Liu—describes details for conjugation of TF AP-1 or control synthetic double-stranded phosphorothioate end-capped DNA aptamers to superparamagnetic iron oxide nanoparticles (SPIONs), gold, or fluorescein for imaging by MRI, transmission electron microscopy studies, and optical methods, respectively. The SPION-enabled MRI results revealed that neuronal AP-1 TF protein levels were elevated in neurons of live mice after amphetamine exposure, as expected, while transgenic mice with neuronal dominant-negative A-FOS mutant protein, which has no binding affinity for the AP-1 sequence, showed a completely null MRI signal. These investigators concluded that further clinical development is warranted.
Shortest aptamer yet?
Also after finishing this blog, an 11-mer aptamer was reported by Nadal et al. to have been obtained by truncation of an aptamer that specifically binds to β-conglutin (Lup an 1), an anaphylactic allergen. The highest affinity was observed with a truncation resulting in a G-quadruplex-containing 11-mer sequence that had an apparent equilibrium dissociation constant (KD) of 1.7 × 10-9 M. This 11-mer sequence was demonstrated to have high specificity for β-conglutin and showed no cross-reactivity to other lupin conglutins (α-, δ-, γ-conglutins) and closely related proteins such as gliadin.
Another Resource
Nucleic Acid Therapeutics can be searched for “aptamers” with other keywords for full-text access to numerous publications on various aspects of RNA or DNA aptamer selection, targeting, etc. and reviews thereof.
Language Trivia
The word “aptamer” is used regardless of what language you happen to be reading or speaking. The same is true for “blog”, although in German someone might instead use “digitale Netztagebücher”. Since even for German speakers that doesn’t exactly flow off the tongue, the latest edition of Duden—the German equivalent of the Oxford English Dictionary, completely unrelated to “dude”—added 5,000 new words, many of them in English, such as “blog” and “flashmob”, according to Anna Sauerbrey, a German, in the New York Times Op-Ed on September 26, 2013.