
Prime Time Editing
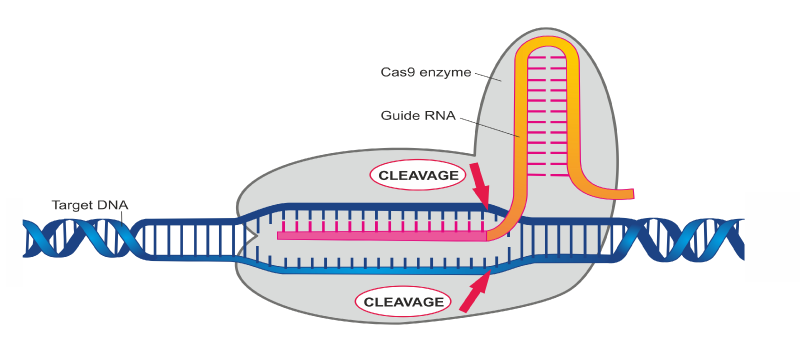
The rapid development of CRISPR gene editing technologies in mammalian systems has massively enhanced the capabilities of modern molecular biology to specifically alter an organism’s DNA. The CRISPR system utilizes components of a natural bacterial immune process. The main enzyme, Cas9, uses a guide RNA to direct it to a site of genomic DNA, where it cuts both strands. The DNA cut is resolved by one of two natural DNA double stranded break processes: non-homologous end joining, or homology directed repair. If homology directed repair is desired, a double stranded DNA repair template must also be introduced with Cas9 and the guide RNA. Through the non-homologous end joining pathway, the CRISPR-Cas9 system is most efficient at generating small insertions or deletions at the cut site. Currently, CRISPR-Cas9 is an actively used and powerful tool in the laboratory space, providing researchers with a new method to affect gene expression in a specific manner. Although it is an impressive new tool, CRISPR-Cas9 does have its limitations. In particular, off-target editing effects and the limited number of efficiently made mutations are major concerns that impede its clinical development.
In October 2019, a group from the Broad Institute described a new version of gene editing, termed prime editing, which circumvents many of the challenges of the CRISPR-Cas9 system. Prime editing has three major components: one, a catalytically impaired Cas9 nickase enzyme that only cuts one strand of DNA; two, a prime editing guide RNA or pegRNA that directs the location of the nick and provides a repair template; and three, a reverse transcriptase enzyme (fused to the Cas9 nickase) that reverse transcribes the pegRNA to make the genetic change. The pegRNA is similar to a guide RNA, but also includes a primer binding site at the 3’ end and a repair template on the 5’ end. TriLink Biotechnologies has the capability to synthesize these long customized pegRNA molecules. Thus, prime editing is a self-contained search and repair system that does not rely on endogenous pathways and has the capability of executing any desired genetic change.
In the Nature article that describes prime editing, authors developed their system in vitro and in yeast before doing the bulk of their work in HEK293T cells. They performed every single nucleotide substitution at 5 different genomic loci, showed 28 successful small insertion and deletion edits, implemented larger changes up to 80 base pairs, and performed combination changes. When they examined off-target effects, they found prime editing to be 4.4x less active at known off-target sites compared to the typical CRISPR system. Transcriptomic analysis showed fewer than 100 genes with gene expression changes greater than 2 fold. The authors examined three pathogenic mutations by first creating and then correcting mutations that cause sickle cell disease and Tay Sachs disease, as well as introducing a prion-protective mutation. The work in HEK293T cells was followed by feasibility studies in three more human cell lines, with notably lower efficiencies. In the final system they tested, the authors showed limited editing efficiency in vivo in embryonic mouse neurons.
Much more work needs to be done on our understanding of this technique and what it can help science accomplish. Outstanding questions could include: do different mutations at the same locus require re-optimization of the system for the same desired edit? Are there genomic rearrangements or alterations occurring at distal sites? What is the efficiency of prime editing pathogenic mutations in a relevant cell context or disease model? Experts believe it will not replace CRISPR-Cas9, but the many variations of these systems can and should be used on a case by case basis. The power, precision, and flexibility of prime editing demonstrated in this paper is impressive. If this technique can succeed clinically, it could correct 89% of known pathogenic variants. Clearly, once better understood, prime editing holds tremendous promise.
Featured product: Custom mRNA Synthesis
Anzalone, A. V. et al. Nature https://doi.org/10.1038/s41586-019-1711-4 (2019)

