
Novel Selection Process Leads to Improved Aptamers for HER2-Positive Cancer
July 2016
Recently, Belgian researchers showed that a combination of next-generation sequencing, a novel method termed “Dubbles”, and the TriLink 40 nucleotide random library led to the identification of potent aptamers that may provide new diagnostic and therapeutic avenues for patients with human epidermal growth factor receptor (HER2) positive breast cancer.
The research led by Marlies Gijs, from the Belgian Nuclear Research Centre, was published last month in Pharmaceuticals.
HER2 is a growth promoting transmembrane protein that is overexpressed in approximately 15-20% of breast cancers and is associated with poor patient prognosis. Current standard of care often includes trastuzumab, a humanized monoclonal antibody targeting the HER2 protein.
“However, only one third of patients respond to trastuzumab therapy and most of the responders eventually relapse. Moreover, resistance to trastuzumab is of major concern and heart failure occurred in 1% to 4% of patients treated with trastuzumab,” the researchers note.
As a surface protein, HER2 is an ideal molecular target, thus the authors sought another avenue for HER2 targeting through the development of aptamers. Aptamers are single stranded, non-coding oligonucleotides typically greater than 70 nt in length and contain primer regions on the 5´ and 3´ ends, flanked by a random region.
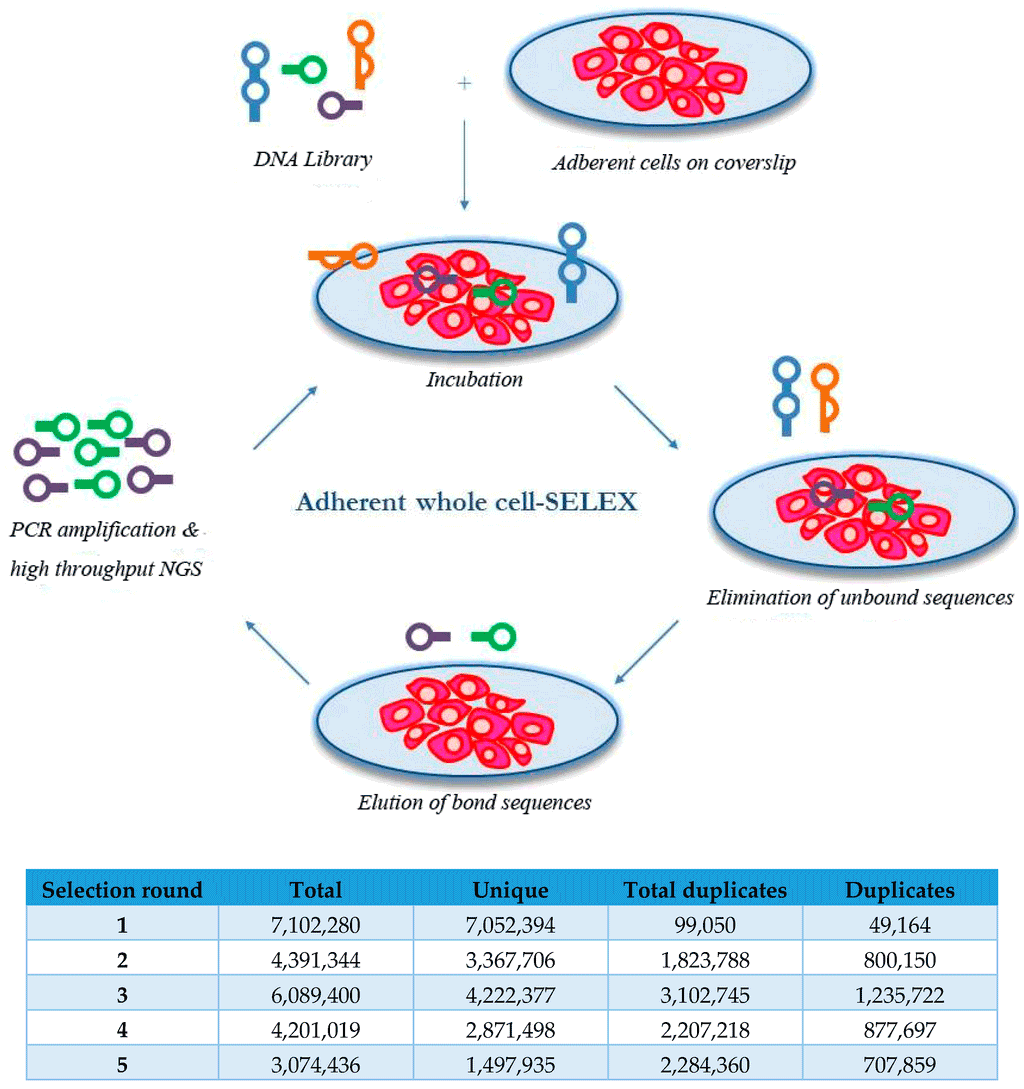
Aptamers are typically identified through systematic evolution of ligands by exponential enrichment (SELEX). This involves the incubation of a large pool of random sequences with a target ligand. Those that do not bind the target are removed and the bound sequences are eluted and amplified by PCR to prepare for subsequent rounds of selection (Figure 1). As this process is repeated, the stringency of the elution conditions is often increased to help identify the tightest-binding sequences.One problem that occurs during the selection process involves the hybridization of the aptamer primer regions to the random region, which leaves the random region unavailable for the target sequence. To circumvent this problem, the researchers used a new method developed by Neoventures Biotechnology Inc. called “Dubbles”.The Dubbles process discourages the hybridization of primer sequences to the complementary random regions through a “snap-freeze” step during amplification. Additionally, at the end of the selection process, the random region is then synthesized without the primer region, leaving the aptamers only ~40 nucleotides in length, which increases synthesis efficiency (and yield) and decreases cost. Using the TriLink 40 nucleotide random library and the Dubbles technique, the authors were able to identify multiple candidate aptamers targeting the HER2 protein in adherent breast cancer cells (Figure 2).

Previously, it could take up to 16 laborious rounds of selection to identify aptamers of high affinity and specificity to the target. However, the use of next-generation sequencing allows for the identification of optimal aptamer sequences with fewer rounds through the ability to count the frequency of each sequence (abundance) and its rate of increase per selection round (enrichment). Here, the authors were able to identify candidate aptamers to HER2 in only five rounds.Furthermore, additional experiments showed that two of these aptamers also abrogated cell growth (Figure 3). One aptamer was internalized by cells overexpressing HER2, making it a potential delivery vehicle for therapeutic payloads.

Though more in vivo research needs to be done to validate the aptamers, this approach may provide another therapeutic avenue for HER2-positive cancers, particularly those who do not respond well to trastuzumab.Featured Product: 40 nucleotide random library. TriLink has utilized their expertise to ensure as close to a 1:1:1:1 ratio of bases as possible during oligo synthesis. Each batch is tested by enzymatic digest to determine base composition and to verify that it falls within rigorous TriLink specifications, ensuring lot-to-lot reproducibility. Please contact us for a quote or to discuss your project.

