
Alternative Splicing is Highly Sensitive to Mutation
August 2016
Even before the nature of genes were known and before Mendel first established the basic rules of heredity through his experiments in pea plants, the relationship between genes and their observable traits has been a major question for scientists.
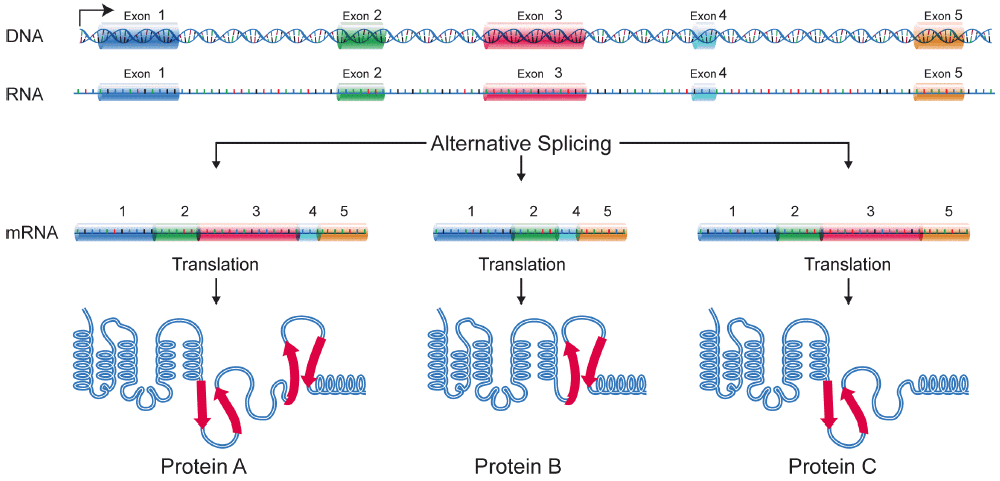
Though it has long since been determined that inheritable traits are attributable to segments of DNA passed down from each parent, one facet of the genotype-phenotype relationship that is still relatively undetermined is the regulation of gene expression through alternative splicing, a post-transcriptional process that alters the inclusion/exclusion of exons (Figure 1).
Though alternative splicing has been of particular interest in recent years because of its link to diseases such as cancer, it is poorly characterized, primarily because of the complexities involved in the research.
However, in a recent issue of Nature Communications, Spanish researchers sought to understand how alternative splicing is regulated by studying the effects of nearly all single and double mutations of the 63 nucleotide FAS/CD95 exon 6. FAS/CD95 is a death receptor and the alternative splicing of exon 6 generates isoforms with opposite roles in apoptosis.
To do this, researchers had TriLink create a 135 nucleotide-long degenerate oligonucleotide library, which included the FAS/CD95 exon 6 sequence. Each position was doped with 1.2% of each of the three non-reference nucleotides to create nearly every possible single and double mutation in the target sequence.
Researchers then used this library to look at the effect of each mutation on gene expression in HeLa cells. Interestingly, they found that 39% (74/189) of single mutations significantly decreased inclusion of exon 6 while 22% (41/189) significantly increased inclusion. 39% (73/189) had no statistically significant effect. Further analysis showed that single nucleotide mutations in 92% (58/63) of the positions affected splicing (Figure 2).

Analysis of 95% of the possible double mutations (16,728/17,577) indicated that although double mutants have similar rates of inclusion, they have a much broader distribution of effects. They also noted that the wild-type sequence is more sensitive to change than the same sequence with one or more mutations (Figure 3).

Finally, they looked at epistasis, which is defined as “the unexpected outcome of combining two or more mutations.” To measure epistasis, they quantified gene expression of a near neutral mutation combined with a second mutation.
They found that 40% (76/189) of all mutations interact with a near-neutral mutation by altering gene expression and 74% (54/73) of near-neutral mutations modify the outcome of at least one mutation. They also found that epistatic interactions are more likely to occur locally, suggesting a synergistic relationship between mutations in local regulatory sequences.
While this work convincingly demonstrates that alternative splicing is highly sensitive to mutation in FAS/CD95 exon 6, how applicable this is across the genome is unclear. Regardless, this work has many important implications for evolutionary biology and disease and it has carved a new path for future research on alternative splicing.
Featured Product: Custom degenerate/wobble site oligonucleotide library. TriLink offers the standard IUPAC wobble options, as well as custom wobbles (randomers). We hand mix our amidites and supports to ensure a proper distribution at each site in your oligonucleotide, may that be 1:1:1:1 or a custom ratio.

