
Detecting and Controlling Protein Expression with mRNA Devices
August 2017
Synthetic biology is a growing field that introduces exogenous, synthesized biological components to influence natural biological systems. Synthetic biology has pioneered many new therapeutic strategies, including gene and protein replacement through the introduction of modified mRNA, DNA, or modified immune or stem cells into an organism.
Beyond our current capabilities, however, is the goal to program cell fate autonomously. This requires rapid detection and response to intracellular protein levels enabling a more precise, sensitive and sophisticated system to regulate cell fate.
Recently, published in Nucleic Acids Research, a group of Japanese researchers set out to design mRNA constructs with improved sensitivity to detect endogenous proteins in human cells, allowing the information to be transmitted to downstream synthetic translational regulatory systems.
"A protein-driven mRNA device that detects a particular target protein and regulates post-transcriptional expression of exogenous genes can be used to build complex and sophisticated gene circuits, because the output protein from the device can serve as the input protein of other circuits," the researchers explain.
The existing techniques to detect specific proteins, such as western blotting, immunostaining, and chemical probes enable protein expression to be characterized, however, these methods often rely on synthetic protein tags and may not be reliable or useful in living cells.
Current protein-responsive RNA devices rely on simultaneous introduction of exogenous RNA-binding proteins that can be difficult to express or exogenous DNA, which can integrate into endogenous DNA and disrupt normal protein production.
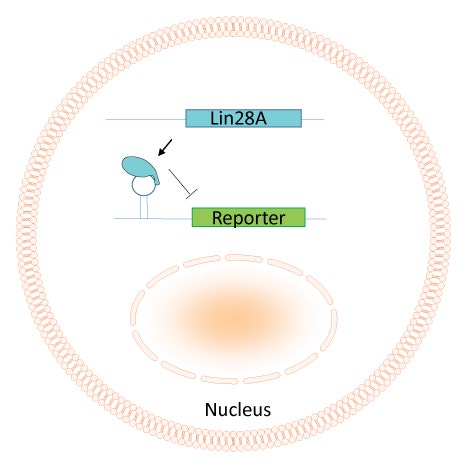
To circumvent these issues, the group developed novel aptamers with RNA binding motifs designed at the 5’ region of the mRNA to protect and stabilize their active conformations in mRNA (Figure 1).
Using TriLinks pseudouridine-5΄-triphosphate (Ψ) and 5-methylcytidine-5΄-triphosphate (m5C), they found that modified RNA interfered with protein binding, which is consistent with previous reports. Thus, the group relied on unmodified mRNA for their protein responsive mRNA devices.
They tested their system with LIN28, a target protein that is critical for controlling cell fate and is highly expressed in human pluripotent stem cells. With this system, they found they were able to reliably and quantitatively detect LIN28, but more importantly could respond to LIN28 expression.
Additionally, the group showed that the mRNA constructs were able to control differentiation of human induced pluripotent stem cells (hiPSCs) through the detection and repression of LIN28. Importantly, they found that this system did not affect expression levels of miRNA that are controlled by LIN28, providing further control of downstream processes.
"…combining [this technology] with current gene regulation technologies, including miRNA responsive switches, RNA inverter modules, cell classifier circuits, ribozyme devices, and two-dimensional tuning (optimized positioning and multiple insertion of RNA motifs in mRNA) will enable us to build more sensitive, precise and increasingly sophisticated systems to program cell fate," the authors conclude.
Featured Products: TriLink has a number of modified bases suitable for mRNA transcription, as well as custom mRNA transcription services. 5-Methoxyuridine-5-Triphosphate (5moU), Pseudouridine-5-triphosphate (Ψ) and 5-methylcytidine-5-triphosphate (m5C). Please contact us to discuss your next project.
Have a question? Visit Ask An Expert.

