
Modified mRNA May Suppress Immune Activation at Multiple Steps of RIG-I Pathway
November 2016
One major hurdle for biological therapies such as messenger RNA (mRNA) is the threat of immune activation. However, over the last decade, scientists have discovered that mRNA containing various modified nucleotides, such as pseudouridine and N-6-methyladenosine can suppress innate immune activation, making mRNA a powerful tool for regenerative medicine, treatment of diseases, and reprogramming of cells. Nevertheless, the detailed mechanisms of how the modified nucleotides evade immune activation are still largely unknown, particularly for lesser studied modified nucleotides.
Last month, researchers from Harvard and Rutgers University published work that sought to uncover the mechanisms that allow these modified nucleotides to evade immune activation through the retinoic acid-inducible gene I (RIG-I)-like pathway, and avoid the downstream interferon-beta (IFN-Β) production. In this study, the researchers used various independent assays, including RIG-I:RNA binding assays, limited trypsin digestion of RIG-I:RNA complexes, and inhibition of virus replication, to try to resolve how RNAs with modified nucleotides evade or suppress innate immune signaling.
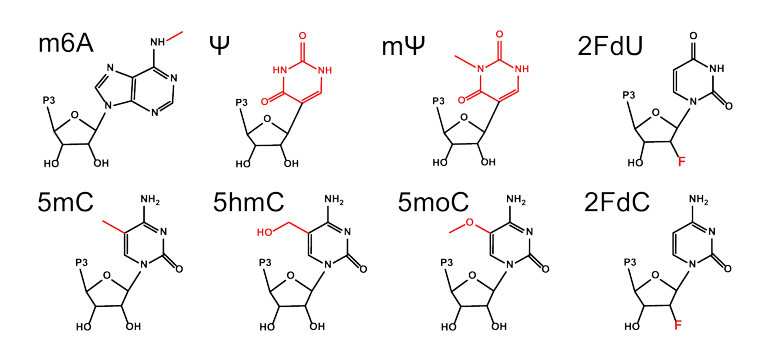
The experiments were based on the polyU/UC RNA domain of hepatitis C virus, a model innate immune activating RNA molecule which was transcribed with canonical nucleotides or with one of eight modified nucleotides, most of which were provided by TriLink (Figure 1). Their initial experiments confirmed previous research indicating that each mRNA with modified nucleotides reduced IFN-Β production compared to unmodified mRNA and verified that IFN-Β production correlated with an antiviral state (Figure 2).

However, subsequent experiments suggested that the modified nucleotides may act through difference mechanisms. For example, through a modified mRNA/RIG-I binding affinity assay, the researchers found that polyU/UC with m6A displayed a much lower affinity to RIG-I, whereas other modified nucleotides showed a little or no difference in binding affinity when compared to canonical nucleotides. Interestingly, they noted that signal transduction intensity didn?t necessarily correlate directly with RIG-I:RNA binding affinity. Together, this suggests that modified nucleotides may be acting to suppress immune activation at different stages in the RIG-I pathway.
To resolve this, the researchers asked whether RNAs containing certain modifications could bind to RIG-I without triggering the conformational changes necessary for downstream signaling. Using a limited trypsin digestion of RIG-I:RNA complexes, they found that the results varied depending on the modification and the presence of ATP or of a nonhydrolyzable ATP analogue, adenylyl-imidodiphosphate (AMP-PNP). This suggests that the modified RNA ligands differ in their propensity to bind the helicase domain of RIG:I, which appears to be driven by the chemical structure of the modified nucleotide, and that they cooperatively bind ATP or the AMP-PNP nucleotide.
"The results [of this study] suggest that RNAs containing modified nucleotides interrupt signaling at early steps of the RIG-I-like innate immune activation pathway and also that nucleotide modifications with similar chemical structures can be organized into classes that suppress or evade innate immune signaling steps [see Figure 3]. These data contribute to defining the molecular basis for innate immune signaling suppression by RNAs containing modified nucleotides. The results have important implications for designing therapeutic RNAs that evade innate immune detection," the researchers concluded.

Featured Product: TriLink offers N-6-methyladenosine (m6A), pseudouridine (Ψ), N-1-methylpseudouridine (mΨ), 5-methylcytidine (5mC), 5-hydroxymethylcytidine (5hmC), 5-methoxycytidine (5moC), other modified nucleotides, and modified EGFP. If you do not see your preferred modified nucleotide or modified mRNA, please contact us for a custom quote.

