
When Timing is Everything. Uncovering the Temporal Dynamics of RNA Using TimeLapse-seq
February 2018
Through the advancement of modern techniques such as RNA sequencing, our knowledge of the transcriptome has increased dramatically and opportunities for new therapies have broadened. However, standard RNA sequencing can only offer a static view of an RNA population and an efficient method to understand the temporal dynamics of RNA is lacking.
A group of researchers from Yale University has developed a new RNA sequencing method that is capable of distinguishing temporal changes in RNA transcription, TimeLapse-seq.
"[Current methods] require large amounts of input sample and extensive handling, and they present challenges when normalizing enrichment and estimating contamination," note the authors.
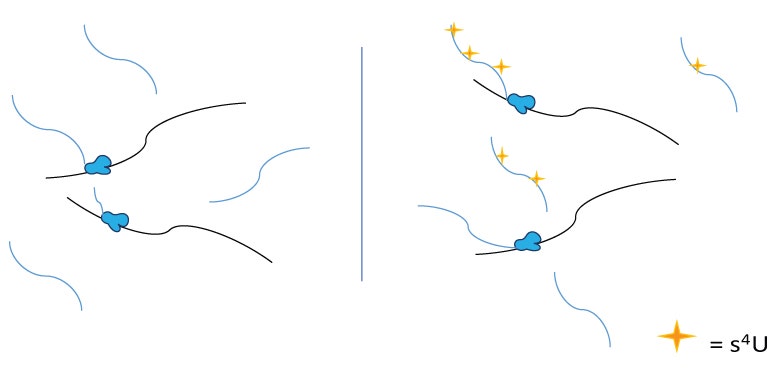
In their article published last month in Nature Methods, the researchers unveiled a new, efficient sequencing technique that starts by tagging new transcripts with the uridine analog 4-thiouridine (s4U: Figure 1).
The 4-thiouridine is then converted into cytidine analogs through oxidative-nucleophilic-aromatic substitution (Figure 2). This leads to U-to-C mutations, and marks new transcripts upon sequencing.

Using this method, the authors were able to monitor global steady-state RNA turnover, as well as distinguish acute transcriptional changes, as demonstrated by analyzing transcription during 1 hour of mild heat shock.
In addition, this technology allows for the identification of isoform-specific transcript dynamics, which may be particularly important since RNA processing has been linked to many diseases.
Specifically, they show that a particular isoform of a gene linked to myelodysplastic syndromes (MDS) has a greater turnover than other isoforms. How this affects MDS is unknown, but the ability to uncover and understand isoform-specific stability may lead to new therapies.
"[Importantly], TimeLapse-seq is broadly applicable to any application compatible with metabolic labeling (e.g., TT-seq). This approach provides a flexible platform to investigate dynamic biological systems," the authors conclude.
Featured Products: 4-thiouridine-5-triphosphate.
TriLink BioTechnologies offers custom chemistry and contract research services that include synthesis of unique phosphoramidites and polyphosphates, including mono-, di- and triphosphates, and other small molecules. If the small molecule your research requires is not commercially available, contact us. Collectively, our R&D team has decades of experience developing and optimizing nucleic acid synthesis schemes.
Interested in sequencing small RNA? The TriLink CleanTag® technology solves one of the most pervasive problems in library preparation for next-generation sequencing (NGS), adapter dimer formation.
Have a question? Visit Ask An Expert.

