- Revisiting a Polymerase Ribozyme for 3’-End Labeling Oligos
- Using a Wide Variety of Modified Nucleotide Triphosphates from TriLink to Demonstrate Versatility of Labeling
 Gerald Joyce. Taken from scripps.edu
Gerald Joyce. Taken from scripps.edu
In September 2016, I wrote a blog featuring a remarkable publication by the Gerald Joyce lab at Scripps Research Institute in La Jolla, CA. The researchers wrote about the in vitro evolution of an RNA catalyst (i.e. ribozyme) that had RNA polymerase activity and could amplify RNA. This purely RNA-based synthetic chemistry, in the complete absence of any proteins, provided further evidence for the feasibility of “RNA world,” a phenomenon first discussed by Walter Gilbert in 1986, who hypothesized the existence of prebiotic era billions of years ago during which life began without DNA or proteins.
This blog post once again spotlights the Joyce lab, but in the context of applying this novel polymerase ribozyme as a means to carry out 3’-end labeling of RNA or DNA with 50 modified nucleotides. I’m pleased to add that many of the requisite modified nucleotide triphosphates were obtained from TriLink! Interested readers can consult this June 2018 publication in Nucleic Acids Research for more details if they wish to supplement the brief overview that will be given here.
Introduction
RNA polymerase ribozymes are in vitro evolved RNA molecules that extend an RNA primer on a complementary RNA template using NTP substrates. Currently, the most advanced RNA polymerase ribozyme is the ‘24-3’ polymerase, which was reported in 2016 by Horning & Joyce to have an extension rate of ~1 nucleotide (nt) per minute, and can operate on most template sequences. Using specially designed templates, the 24-3 polymerase can generally be limited to the addition of only a single modified nucleotide, thus enabling efficient 3’-end labeling of a target RNA or DNA using various NTP and dNTP analogs shown here in red.
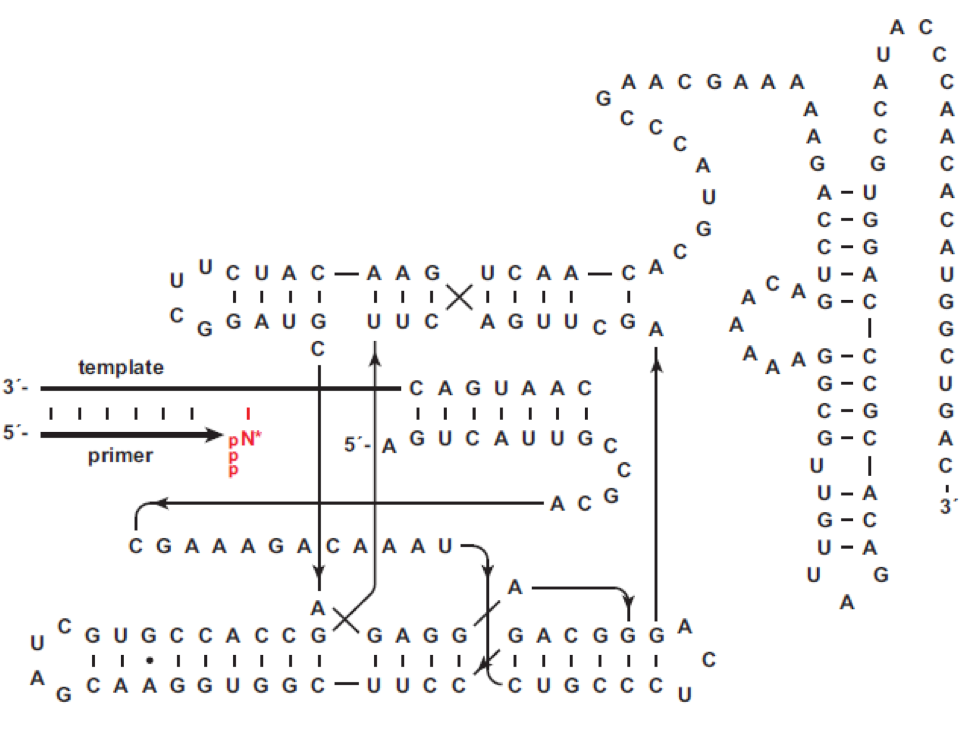 Taken from Joyce and coworkers Nucleic Acids Research 2018
Taken from Joyce and coworkers Nucleic Acids Research 2018
The highly structured 24-3 polymerase ribozyme, which is depicted here in 2D, contains 180 nt. The ribozyme also has a short “tag” sequence (5’ GUCAUUG 3’) at the 5’ end of the polymerase that is complementary to a sequence (3’ CAGUAAC 5’) at the 5’ end of the template. Besides this feature, the template sequence is not constrained. The primer, which corresponds to the template nucleic acid, binds to the template through Watson–Crick pairing and is extended by the polymerase to achieve 3’-end labeling.
Although the 24-3 polymerase ribozyme can add multiple successive NTPs to the 3’ end of a template-bound primer, the reaction can mostly be restricted to the addition of a single residue by choosing an appropriate template and providing only one of the four nucleobase substrates. By way of example and as shown here, four templates were constructed, each with a different templating nucleotide (red) at the first position of primer extension, followed by several non-complementary nucleotides. Together, this set of templates enables the testing of triphosphate analogs containing each of the four nucleobases.
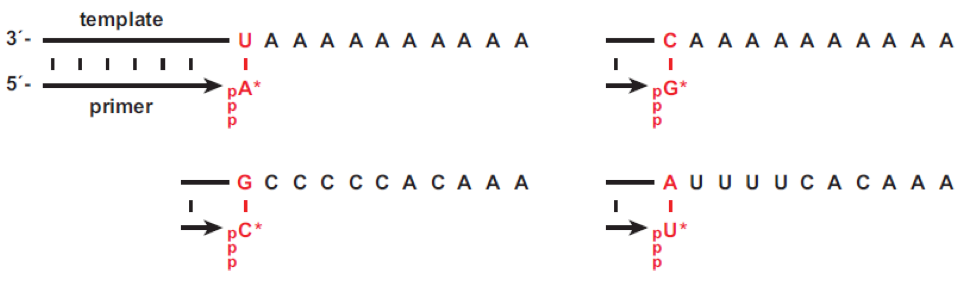 Taken from Joyce and coworkers Nucleic Acids Research 2018
Taken from Joyce and coworkers Nucleic Acids Research 2018
Exemplary Results
Although a great variety of functionalized nucleotides can be prepared by chemical synthesis, this study by Joyce and coworkers focuses on commercially available nucleotide triphosphate analogs, such as sugar, nucleobase, and backbone modifications, in order to demonstrate the general utility of the approach. Fifty different analogs were tested in a reaction employing a 0.8 μM RNA (or DNA) primer with the following sequence: 5’ UUGCUACUACACGAC 3’ (or corresponding DNA sequence), together with 1 μM ribozyme and 1 μM RNA template. The reactions were carried out in the presence of 200 mM MgCl2 and 0.5 mM NTP analog at pH 8.3 at 17◦C for 1 h. Yields by PAGE ranged from 11% to 89% overall, with 84% to 89% yield for five of the analogs.
The exemplary results tabulated here highlight the versatility of 24-3 polymerase ribozyme toward incorporation of NTP analogs with very diverse molecular structures that provide different types of functionality.
Exemplary NTP Analogs (TriLink) and Incorporation Yield by the 24-3 Polymerase Ribozyme
| NTP analog | Yield % | NTP analog | Yield % |
| N6-methyl-2-amino-ATP | 49 | 2’-amino-dATP | 85 |
| 7-propargylamino-dGTP | 89 | 2’-amino-dGTP | 70 |
| biotin-16-aminoallyl-dUTP | 28 | 2’-amino-dCTP | 80 |
| Pseudo-UTP | 66 | 2’-amino-dUTP | 50 |
| α-thio-ATP | 70 | 5-aminoallyl-CTP | 67 |
| α-thio-GTP | 86 | Cy5-aminoallyl-CTP | 47 |
| α-thio-CTP | 84 | 5-formyl-CTP | 85 |
| α-thio-UTP | 11 | 5-formyl-UTP | 58 |
| thieno-GTP | 50 | 1-borano-dGTP | 37 |
| thieno-UTP | 25 | 1-borano-dCTP | 12 |
For instance, N6-methyl-2-amino-ATP is a member of the diaminopurines that are discussed elsewhere, while pseudo-UTP shown below is an isomer of UTP that is now widely used in modified mRNAs.

In a TriLink white paper by Paul and Yee titled PCR incorporation of modified dNTPs: the substrate properties of biotinylated dNTPs, it is noted that the high affinity of streptavidin for the biotin ligand is one of the strongest and most widely utilized interactions in biology. The strength and specificity of this interaction has been exploited in many biological applications, including secondary label introduction and affinity isolation. While there are various length linkers that have been employed for attachment of biotin to the nucleotide, the relatively long biotin-16-aminoallyl-2'-dUTP used for incorporation by 24-3 polymerase ribozyme is often preferred.
Modification of the 3’-end by incorporation of 7-propargylamino-dGTP, 2’-amino dNTPs or 5-aminoallyl-CTP provides a reactive primary amine group as a versatile “chemical handle” to attach virtually any type of moiety that is needed for an application, whether that be a detectable label or synthetic peptide. The α-thio-NTPs (aka 1-thio-NTPs) and 1-borano-dNTPs demonstrate that these phosphate modifications are compatible with the 23-3 polymerase ribozyme.


5-Formyl-nucleotides provide a reactive formyl (i.e. -CHO) group for conjugation reactions with, for example, hydroxylamine-functionalized labels of the type reported elsewhere. The incorporation of thieno-NTPs is interesting because of the inherent fluorescent properties of this relatively new class of analogs offered by TriLink, which can be read about at this link.
Concluding Comments
According to the aforementioned Joyce publication, the simple, one-step installation of a fluorophore or affinity probe using the 24-3 polymerase ribozyme is likely to have broad application, offering an attractive alternative to 3’-end labeling using a polymerase protein such as poly(A) polymerase or terminal transferase. These polymerase proteins operate in the template-independent manner, and thus result in multiple successive additions, unless the NTP analog itself is a chain terminator.
As usual, your comments are welcomed.








