- Circular RNA (circRNA) Formation Serendipitously Discovered in 1991
- Next-Generation Sequencing Reveals circRNA to be Ubiquitous
- circRNA can Function as MicroRNA ‘Sponges’ to Regulate Gene Expression
There’s something seductively simple—and curious—about circles, which are unique in having no beginning or end, unlike most other things. On a less philosophical plane, thinking about circles conjures up incongruent memories of delicious doughnuts and geometric definitions from my youthful days going to the neighborhood bakery and diligently taking notes in my high school geometry class, respectively.

Having said this, you might now appreciate why I was immediately captivated when I recently read about the existence of circular RNA (circRNA). More importantly, I was immediately puzzled over why Mother Nature would want RNA to evolve in this way, when virtually all other eukaryotic RNA (and DNA) is linear?
I decided to research curiously circRNA to learn the answer to this puzzlement, and in so doing found a wealth of interesting molecular biology, much of which was blogworthy. For lack of space here I’ve selected and made into snippets some of these finds that were most interesting—at least to me.
Serendipitous Discovery of circRNA
I’m always interested in the seminal discovery aspects of science, and when researching this for circRNA, I learned that these curious molecular “creatures” were first reported in 1979 by Hsu & Coca-Prados in an article entitled Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. I spent considerable time trying to get an image from that publication but was unsuccessful. However, I was able to find an analogous image published in 1990 by Matsumoto et al. for single-stranded circRNA from yeast, as shown below. Matsumoto noted they needed strong denaturing conditions to disrupt this molecule’s extensive intramolecular H-bonding.
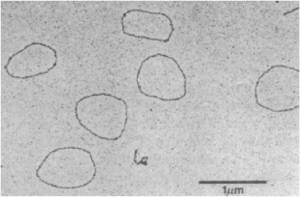 Electron micrograph of ~3,000-nt circular 20S RNA from yeast. Taken from Matsumoto et al. (1990) PNAS.
Electron micrograph of ~3,000-nt circular 20S RNA from yeast. Taken from Matsumoto et al. (1990) PNAS.
Interestingly, one year later, in 1991, uber-famous Bert Vogelstein and coworkers unexpectedly found “scrambled transcripts” present at only l/l000th the concentration of normally spliced linear transcripts of a candidate tumor suppressor gene they were studying. They proposed a mechanism for formation of these rare, scrambled, or “abnormally spliced transcripts” that involved generation of circRNA. This “serendipitous discovery,” as they described it, thus provided indirect evidence for how RNA circularization can occur. But are they only a rare mistake of biochemistry or new species of functional RNA with an evolutionary purpose?
The answer, according to a review by Qu et al., is that until 2010 few circRNAs had been discovered, and research into circRNA biogenesis was minimal. However, with the development of high-throughput sequencing technology and computational analysis, thousands of circRNAs have been discovered across species from simple Archaea (single-cell microorganisms without a nucleus) to high-complexity humans. In fact, expression of some circRNAs is >10-fold higher than those of their canonical linear transcripts of the same genes!
I’ll refrain from trying to summarize the currently known or proposed mechanisms for biogenesis of circRNA because there are quite a few, and are not simple to describe without a lot of jargon, so please see Qu et al. if you’re interested.
Biological Functions of circRNA
‘Sponges’ for microRNA: Remarkably, the first compelling functional analysis of naturally expressed circRNA wasn’t reported until 2013, when Hansen et al. in Denmark did so in venerable Nature magazine. According to these investigators, microRNAs (miRNAs), regulators of gene expression, can be affected by the presence of miRNA ‘sponge’ transcripts in a manner formally analogous to how synthetic antisense modified oligos are used to target miRNA (aka antagomirs).
They showed that a circRNA, which they had previously identified as being highly expressed in human and mouse brain, acts as a sponge for a specific microRNA (miR), namely miR-7. They named this circular transcript ciRS-7 (circular RNA sponge for miR-7). The sponge metaphor is apropos in view of the fact that ciRS-7 contains more than 70 target (i.e., binding) sites, as depicted below.
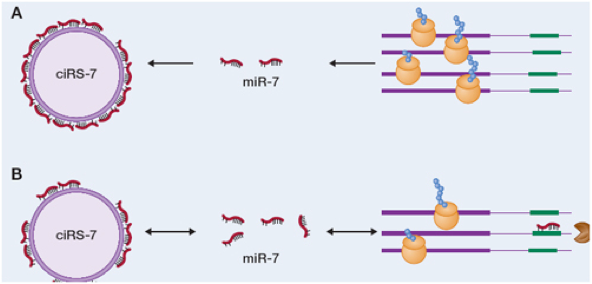 Taken from arraystar.com
Taken from arraystar.com
Moreover, ciRS-7 strongly suppresses miR-7 activity, resulting in increased levels of miR-7 targets. In the mouse brain, they observed overlapping co-expression of ciRS-7 and miR-7, particularly in neocortical and hippocampal neurons, suggesting to them a high degree of endogenous interaction.
Regulation of Alternative Splicing or Transcription: See Qu et al. for details.
Regulation of Expression of Parental Gene: See Qu et al. for details.
Participation in the Carcinogenesis and Malignancy of Cancer: In a review by Zhao & Shen in 2015, they opine on the specific role of circRNAs in a wide variety of cancers and in regulating the biological behavior of cancers via miR-7 (see above) or miR-138 regulation. Furthermore, circRNA, together with its gene silencing ability, also play a role in RNA interference (RNAi) therapy by binding to target RNAs, which provides a novel perspective in cancer treatment.
In conclusion, I hope that you found circRNA to be as curiously captivating as I did. I encourage you to share your thoughts as comments here.
Postscript
For the sake of completeness, and to avoid being misleading, there’s a wealth of information on the various types and function of circRNA, as well as circular extrachromosomal DNA, which is common in prokaryotes but restricted in type in eukaryotes, notably mitochondrial DNA.

Similar to my previous blog post about a high school student’s PCR experiment being carried out on the International Space Station, I recently read about another mindboggling high school student who was recognized for a science project that involves a level of knowledge previously associated with doctoral or post-doctoral education. Matthew Bauer, a sophomore at The Davidson Academy, won the 49th Annual Western Nevada Regional Science and Engineering Fair with his project titled Quantifying Levels of Circularized RNA from RNA Sequencing Data. His project is explained at this link, which also provides a video clip. Kudos to Matthew!






