- DNA isn’t What it Used To Be!
- Scripps Team Led by Floyd Romesberg Demonstrates Six-Base DNA Replication in Living Bacteria
- Are We On Our Way to Semi-synthetic Life-forms? With Vast Potential—But at What Risks?
Doing the Impossible
Seemingly everyone these days knows about DNA and how seemingly simple pairings of A-T and G-C encode all life forms. Now, a team led by Floyd Romesberg, a biological chemist at the Scripps Research Institute in San Diego, California, has created a synthetic base pair, which you can simply think of as X-Y, to produce artificial DNA that replicates with six bases! Click here to read more about Romesberg’s findings that were reported last month in Nature.
 Prof. Floyd E. Romesberg, Department of Chemistry, The Scripps Research Institute, La Jolla, California (taken from utsandiego.com via Bing Images).
Prof. Floyd E. Romesberg, Department of Chemistry, The Scripps Research Institute, La Jolla, California (taken from utsandiego.com via Bing Images).
Expanding the genetic code of DNA has been pursued for decades. Romesberg’s achievement, however, has set the scientific world—and news media—a buzz because his version of expanded DNA (eDNA) actually replicates, setting the stage for future biosynthetic engineering aimed at microbes having correspondingly expanded RNA (eRNA) and—here’s the punchline—greatly expanded protein (e-protein) diversity.
Whereas natural proteins are comprised of 20 amino acids encoded by four-base RNA/DNA, Romesberg’s e-proteins could be comprised of 172 (!) amino acids—both natural and mostly artificial—as the result of more triplet codons derived from six-base eRNA/eDNA.
As listed in the graphic below taken from The Wall Street Journal, this stunning achievement in DNA manipulation research is a very big deal in view of the large number of potential applications it enables, covering virtually the entire spectrum of biotechnology and health.
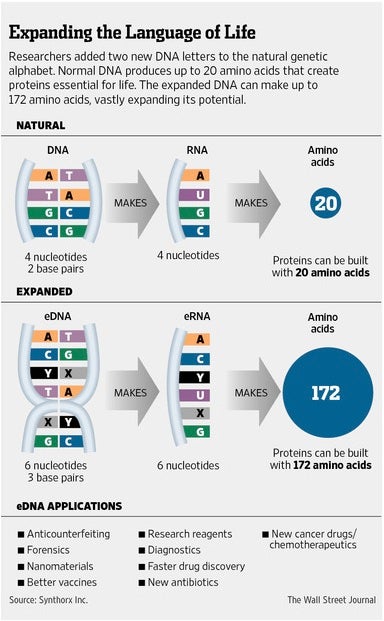
The remainder of this post provides a bit of technical detail and, perhaps more interestingly, some reported opinions that are decidedly positive or—not surprisingly—strongly negative because of concerns for “unintended consequences” of the sort that society has experienced in the past.
Look Folks, no Watson-Crick H-bonds!
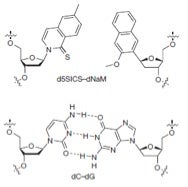 Structure and paired orientation of the unnatural X-Y base pair compared to natural C-G base pairing (taken from Romesberg and coworkers Nature 2014).
Structure and paired orientation of the unnatural X-Y base pair compared to natural C-G base pairing (taken from Romesberg and coworkers Nature 2014).
What immediately struck me as being quite unexpected—if not amazing—about the unnatural X-Y base pair was the absence of Watson-Crick H-bonding commonly associated with specific A-T and G-C base pairing. Instead, these X and Y bases—with actual abbreviations d5SICS and dNaM—have relatively simple bicyclic aromatic rings and minimalistic substituents. My assumption is that these hydrophobic moieties—which aren’t even actual bases (!)—have very specific complementary geometry and “pi-stacking” interactions with hydrophobic moieties in flanking A-T and/or G-C base pairs.
Readers interested in technical details underlying this feat—of which there are many—will need to read the entire report. However, some of the “tricks” used by the Romesberg team are worth mentioning here, admittedly in over-simplified terms for the sake of brevity.
- Their earlier results indicated that passive diffusion of unnatural nucleosides into microbes was possible but subsequent conversion to unnatural nucleotide triphosphates that are required for DNA synthesis was inefficient.
- That problem was cleverly finessed by “borrowing” a suitable nucleotide triphosphate transporter (NTT) from a certain eukaryotic marine phytoplankton. They also had to include a couple of chemicals in growth media for this NTT to adequately function.
- Conventional molecular biology was used to prepare a circular double-stranded plasmid having X-Y at a specific locus for transfection into E. coli bacteria to determine if bacterial DNA polymerase would replicate X and Y when “fed” the triphosphate forms of X and Y (dXTP and dYTP).
After 15 hours of growth, which represented 24 doublings or 2 X 107 amplification of the initial X-Y plasmid, analysis confirmed what had been hoped—namely, the unnatural X-Y base pair was retained during replication. Voilà!
They also investigated resistance of X-Y base pairs to E. coli DNA excision-repair processes after reaching stationary phase, and found X-Y to be quite stable: 45% and 15% retention (toward replacement by A-T) after days 3 and 6, respectively.
Here’s what Romesberg and coworkers opined in their concluding remarks:
“In the future, this organism, or a variant with the [unnatural base pairs>
incorporated at other episomal or chromosomal loci, should provide a synthetic biology platform to orthogonally re-engineer cells, with applications ranging from site-specific labeling of nucleic acids in living cells to the construction of orthogonal transcription networks and eventually the production and evolution of proteins with multiple, different unnatural amino acids.”
At the risk of belittling this major milestone, there is clearly much more to do in order to extend—pun intended—X and Y into unnatural eRNA and, eventually, unnatural e-proteins prophetically imagined in the above graphic. Obviously there are many challenges ahead, but as the saying goes "every journey begins with a single step,” and in my opinion Romesberg’s team has taken a huge leap forward.
eDNA opens a world of possibilities in terms of unique products for companies synthesizing nucleotides. TriLink would certainly like to add all sorts of future dXTP and dYTP (and ribo versions) to their already extensive offering of modified nucleotides. Hopefully, that won’t be too far in the future. Time will tell.
Laudatory Views and Some Expression of Concern
My quickie survey of quotations in various stories reporting this work by Romesberg’s team, which incidentally included scientists at enzyme-purveyor New England Biolabs, indicated mostly high praise.
For example, a May 7th NY Times story by Andrew Pollack quotes Eric T. Kool, a professor of chemistry at Stanford who is also doing research in the area, as saying “it took some clever problem-solving to get where they got,” adding “it is clear that the day is coming that we’ll have stably replicating unnatural genetic structures.”
A May 9th editorial by Robert F. Service in venerable Science magazine quotes Ross Thyer, a molecular biologist at the University of Texas, Austin, as saying that "this is an amazing enabling technology," and that the feat opens the way to a universe of new proteins—a vastly more diverse menu of proteins with a wide variety of new chemical functions, such as medicines better able to survive in the body and protein-based materials that assemble themselves.
Despite the resoundingly positive feedback, some concern about eDNA has been voiced. Jim Thomas of the ETC Group, a Canadian advocacy organization, said in an email, “The arrival of this unprecedented ‘alien’ life form could in time have far-reaching ethical, legal and regulatory implications. While synthetic biologists invent new ways to monkey with the fundamentals of life, governments haven’t even been able to cobble together the basics of oversight, assessment or regulation for this surging field.”
As for scary possibilities such as creating an unnatural dangerous organism, the editorial in Science says that creating synthetic “superbacteria” might sound ominous, but Kool thinks the risks are low. "These organisms cannot survive outside the laboratory," Kool is quoted saying. "Personally, I think it's a less dangerous way to modify DNA than existing genetic engineering,’ Kool is again quoted as saying.
I’m in the camp of carrying on with this line of research as long as the unnatural base pairs have to be chemically synthesized and “fed” to host organisms in a legitimate lab or manufacturing facility, thus having virtually zero possibility of unintended growth and function.
What do you think?
As usual, your comments are welcomed.






