- CRISPR Craze Continues Led in Part by Wunderkind Feng Zhang
- Nonspecific CRISPR-C2c2 “Collateral” Cutting Channeled into Diagnostics
- Turning Biochemical “Lemons to Lemonade”
This blog post is about a new and powerful diagnostic approach based on CRISPR, which I’ll get to below. But first I’d like to point out several reasons why this is an especially interesting development:
 Taken from spyhollywood.com
Taken from spyhollywood.comAny new method using CRISPR is more “sizzle” for this “super-hot” technology
- Feng Zhang, the 32-year-old author of this publication, is regarded as a wunderkind
- My past blog post on CRISPR-C2c2 “collateral” cutting noted possibilities for turning “lemons into lemonade”
- Such “lemonade” has emerged as a diagnostic cleverly acronymized as SHERLOCK
So, without further ado, here’s a brief recap of CRISPR-C2c2 as an intro to SHERLOCK (think Holmes), followed by why this acronym is apropos for a diagnostic method that magnifies detection, in this case Zika virus, about which I’ve previously commented.
What’s CRISPR-C2c2?
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (CRISPR-Cas) adaptive immune systems in microbes contain endonucleases that can be leveraged for CRISPR-based gene editing using targeted CRISPR RNAs (crRNAs), as I’ve outlined in a previous blog post. While such Cas enzymes target DNA, others target RNA and function as RNA-guided RNases, which was reported by Feng Zhang and collaborators in Science last year in an article titled C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector.
My blog post about that C2c2 publication provides more information, but the main point related to the present blog post about diagnostics is that after recognition of its RNA target, activated C2c2 engages in nonspecific (aka “collateral”) cleavage of nearby non-targeted RNAs. This post-cleavage non-specificity can be transformed, if you will, from something “bad” to something “good” in terms of mechanisms, which I described as being akin to converting ‘lemons into lemonade’. You’ll now see how SHERLOCK is such ‘lemonade’ concocted by Zhang and coworkers.
What is SHERLOCK?
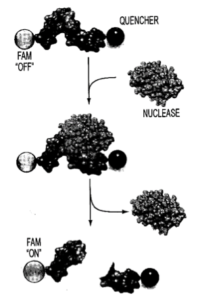 Taken from US 20140199245 A1
Taken from US 20140199245 A1
Before answering this question, and not to confuse you, I need to first mention that the nuclease originally called C2c2 was renamed Cas13a to be more systematic, and will be referred to herein as such. Having said that, Cas13a can be reprogrammed with crRNAs to provide a platform for specific RNA sensing. Molecular recognition of an intended RNA target by crRNA in vitro serves as a sequence-specific “trigger” to activate Cas13a’s nonspecific, “collateral” cleavage of labeled RNA reporters. Cleavage of these fluorescently labeled, internally quenched RNAs leads to unquenching and fluorescent light emission for detection, as depicted below for a generic RNase.
Now that you have a sense of how CRISPR-Cas13a can generate a signal, you’re probably asking ‘What is SHERLOCK?’ It’s an acronym coined by a large team lead by CRISPR genome-editing pioneer Feng Zhang and bioengineer James Collins, both from the Broad Institute. SHERLOCK stands for High-sensitivity Enzymatic Reporter UnLOCKing, which is a recently published new method for sequence-specific detection of DNA or RNA in any sample of interest. This general utility represents a major advance in CRISPR-based diagnostics.
 Feng Zhang. Taken from be.mit.edu // James Collins. Taken from achetron.com
Feng Zhang. Taken from be.mit.edu // James Collins. Taken from achetron.com
As depicted below, sample prep workflows allow for input of either double-stranded DNA (dsDNA) or RNA, which are first amplified by recombinase polymerase amplification (RPA) directly or as cDNA, after reverse transcription (RT-RPA), respectively. In either case, further amplification utilizes T7 transcription into RNA (blue = target RNA; yellow = non-target RNA). Following cleavage of target RNA by Cas13a-crRNA, a commercially available “cleavage reporter” (i.e. internally quenched) RNA undergoes “collateral” cleavage to generate a reporter signal (i.e. fluorescence; pictured below) for real-time detection of the target.

An advantage of RPA is that it is performed isothermally, which allows simplification of the detection device by eliminating the need for a thermal cycler, thus lending to incorporation into small, portable point-of-care (POC) systems (about which I previously commented several times). The abovementioned RPA/T7 sample prep methodology was shown to have attomolar (aM, 10−18 mol/L) sensitivity in model systems, and was next investigated for sensitivity and specificity of virus detection, as well as simplification of reagent format.
Zhang, Collins & coworkers constructed lentiviruses harboring genome fragments of either Zika virus (ZIKV) or the related flavivirus Dengue (DENV), and showed that SHERLOCK detected viral particles down to 2 aM could discriminate between ZIKV and DENV. To explore the potential for POC use of SHERLOCK with paper-spotting and lyophilization (aka freeze-drying), they first demonstrated that Cas13a-crRNA complexes lyophilized and subsequently rehydrated could detect 20 femtomolar (fM, 10−15 mol/L) non-amplified RNA, and that target detection was also possible on glass fiber paper.
The other components of SHERLOCK, namely the RPA reagents and T7 polymerase, were already known to be amenable to freeze-drying and storage at ambient temperatures. In combination, freeze-drying and paper-spotting the Cas13a detection reaction resulted in comparable levels of sensitive detection of RNA as aqueous reactions. Although paper-spotting and lyophilization slightly reduced the absolute signal of the readout, SHERLOCK could readily detect mock ZIKV virus at concentrations as low as 20 aM. Most importantly, SHERLOCK detected ZIKV in clinical isolates (ZIKV RNA extracted from patient serum or urine samples and reverse transcribed into cDNA) could be detected at concentrations down to 1.25 × 103 copies/mL (2.1 aM), as verified by qPCR.
Concluding Comments
If you reflect upon the schematic for SHERLOCK, you’ll note that the input can be either DNA or RNA, which get amplified to produce many copies of RNA that serve as substrates for cleavage by Cas13a-crRNA, thus inducing collateral cleavage of reporter RNA to produce a detectable signal. Lest you think SHERLOCK is too costly to be practical, its developers provide a detailed cost accounting that estimates $0.61 per test, which you’re welcome to compare with your cost of conventional qPCR. I’m quite sure that will lead you to concur with me that $0.61 per test is relatively inexpensive.
If you’re questioning whether SHERLOCK is generally applicable, I urge you to read Zhang, Collins & coworkers in its entirety to learn more about SHERLOCK’s proven ability to detect and distinguish (1) various bacterial pathogens, (2) single-base cancer mutations in cell-free DNA, and (3) health-related single-nucleotide polymorphisms (SNPs) benchmarked against 23andMe genotyping data as the gold standard of these SNPs.
In an article in Science about SHERLOCK, Harvard University’s George Church (who I’ve proclaimed is The Most Interesting Scientist in the World and is the co-founded of a CRISPR therapeutics company), sums up his reaction in one word: ‘Wow.’ I agree!
The article concludes with Collins saying the Broad is now ‘aggressively exploring’ how to commercialize SHERLOCK and may launch a startup company. But before a diagnostic comes to market, it must pass muster at regulatory agencies such as the U.S. Food and Drug Administration. I’m betting is does.
I welcome your sharing any thoughts or comments about this new CRISPR-based diagnostic method.






