- Ned Seeman’s Beer-Assisted “Epiphany” for DNA Self-Assembly Has Led to DNA Nanotechnology
- Creating DNA Origami Is the DNA-Equivalent of the Japanese Art of Paper Folding
- You’ll Be Amazed at What Can Now Be Made from DNA Origami
I first came to know Nadrian “Ned” Seeman by his call-in to me many years ago at Applied Biosystems when he wanted my help in synthesizing highly self-complementary DNA oligos, which can sometimes be challenging. He told me about his interest in using these oligos to study unusual DNA structures related to naturally occurring Holliday junctions. I was intrigued, agreed to help, and we met shortly thereafter at a Gordon Conference on nucleic acids. Little did I know then that Ned’s seemingly quirky-to-me ideas would eventually lead to an entirely new class of self-assembling macromolecules now referred to as DNA nanostructures.
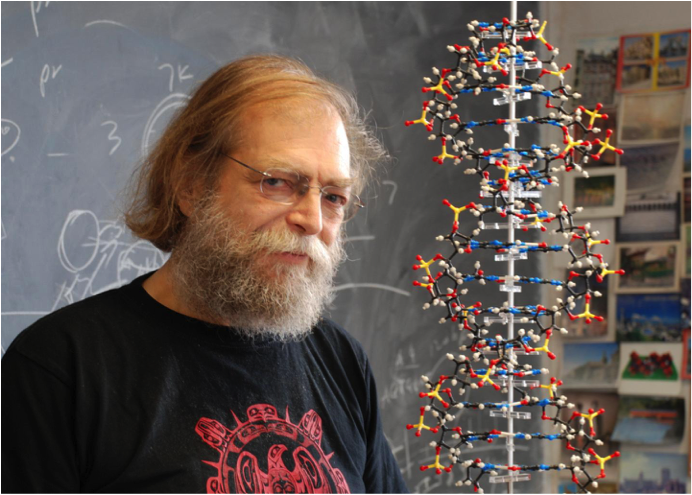 Ned Seeman. Taken from eurekalert.org
Ned Seeman. Taken from eurekalert.org
This video link will take you to Ned Seeman’s personal recollection of how, back then, his drinking beer to better muse about imaginary 6-armed junctions of DNA—instead of Holliday’s 4-armed version—led to Ned’s “epiphany,” in which he envisaged self-assembly of his hypothetical structures to be like Escher’s “Flying Fish,” woodcut print shown here.
 Flying Fish, by MC Escher, 1955. Taken from postcrossing.com
Flying Fish, by MC Escher, 1955. Taken from postcrossing.com
 Taken from dreamstime.com
Taken from dreamstime.com
For reasons that you’ll appreciate by reading on, Ned’s use of simple A/T and G/C base pairing (i.e. hybridization) for the design of elaborate self-assembling DNA nanostructures has paved the way for the creation of some amazing constructs involving DNA folding. Because this process is akin to creating origami sculptures by paper folding, the phrase DNA origami has come into usage.
Having introduced you to Ned and his fathering of the field of DNA nanostructures, I think his 1991 publication in venerable Nature magazine titled Synthesis from DNA of a Molecule with the Connectivity of a Cube catapulted his ideas into high visibility by the scientific community. Interested readers should consult this now classic paper for details on how this was achieved, but for now the below model of such a cube, taken from his lab’s visually entertaining website, reveals the overall cubic shape.
In this representation of a DNA cube, the DNA backbones are shown in red (front), green (right), yellow (back), magenta (left), cyan (top) and dark blue (bottom). Each nucleotide is represented by a single colored dot for the backbone and a single white dot for the base. Each edge of the cube is a piece of double helical DNA, containing two turns of the double helix. Ned’s 1991 Nature paper reporting “the first construction of a closed polyhedral object from DNA” concludes with the sentence below, which in retrospect is both prescient and an understatement of what would then follow his work:
“The synthesis of this object establishes that it may be feasible to make larger and more complex objects.”
Smile!
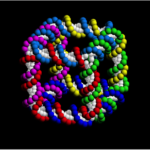
Fast forward nearly 30 years and let me take you to some exemplary “larger and more complex objects” selected from a lengthy, comprehensive 2017 review by Hong et al. titled DNA Origami: Scaffolds for Creating Higher Order Structures. According to this review, Paul Rothemund at California Institute of Technology was the first to report the DNA origami technique in a 2006 paper in Nature. The self-assembly process for a DNA “smiley face” is depicted here.
The steps involved are: (A) a target shape is first approximated with parallel cylinders that are (B) tightly aligned in parallel and that (C) utilize DNA helices. (D) An example of a DNA origami scaffold with “staple” strand routing—use your screen zoom to see these better; the dark blue strand is the “scaffold”, and the staple strands are shown in various colors. (E) The scaffold and the staples (with a large excess of the staples) are mixed, and the designed structures self-assemble with a cooling temperature ramp in a salted buffer. (F) The atomic force microscopy (AFM) image of an assembled DNA origami with a smiley face pattern.
Hong et al. say that rather than using a chemically synthesized strand, it is much cheaper and convenient to use a strand of single-stranded DNA from a viral genome as the scaffold because these genomes are thousands of nucleotides long and originate from a bacteriophage virus with known sequences. The genome of the M13mp18 virus is the most widely used scaffold for assembling DNA origami structures. The scaffold and the staple strands are mixed in buffer and slowly annealed from 90 °C to room temperature, allowing each short staple to find its unique position on the scaffold strand and to ultimately form the desired structure. Voilà!
3D Too!
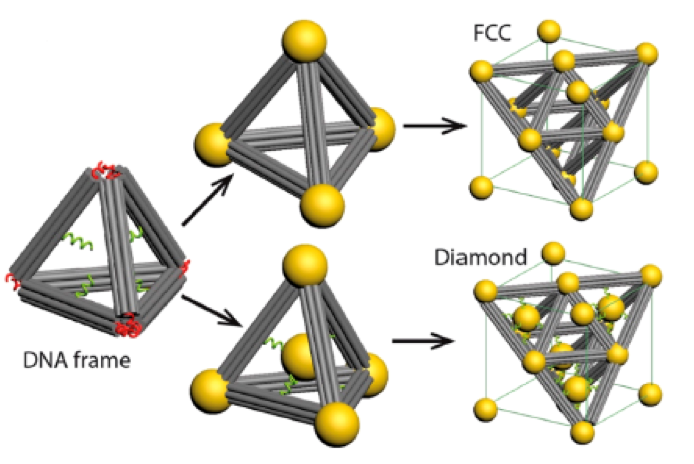
DNA origami provides support with a well-defined geometry for the 3D lattice of nanoparticles’ growth, as demonstrated in 2016 by Liu et al., who designed a tetrahedron-shaped DNA origami with gold nanoparticles attached to the four vertices as shown here. These nanoparticles then acted as connectors to link the tetrahedron origami-nanoparticle units into a well-ordered face-centered cubic (FCC) lattice structure. If another gold nanoparticle was positioned inside the tetrahedron-shaped origami structure, the final lattice structure achieved would be in the shape of a diamond.

In 2017, Ong et al. published in Nature use of LEGO-like DNA “bricks” with 13-nt binding domains to self-assemble 0.1–1-gigadalton 3D DNA nanostructures from tens of thousands of unique components. To put 1 gigadalton (1 GDa) in perspective, a carbon-12 atom has a mass of 12 Da, a molecule of acetylsalicylic acid (aspirin) has a mass of 180 Da, and Titin—the largest known protein—has a mass of 3,000,000 Da or 3 megadaltons (3 MDa). Giga is a short-form of billion, so 1 GDa is 1 x 109 Da. By my estimate, that’s ~2,000,000 base pairs which predictably self-assemble!
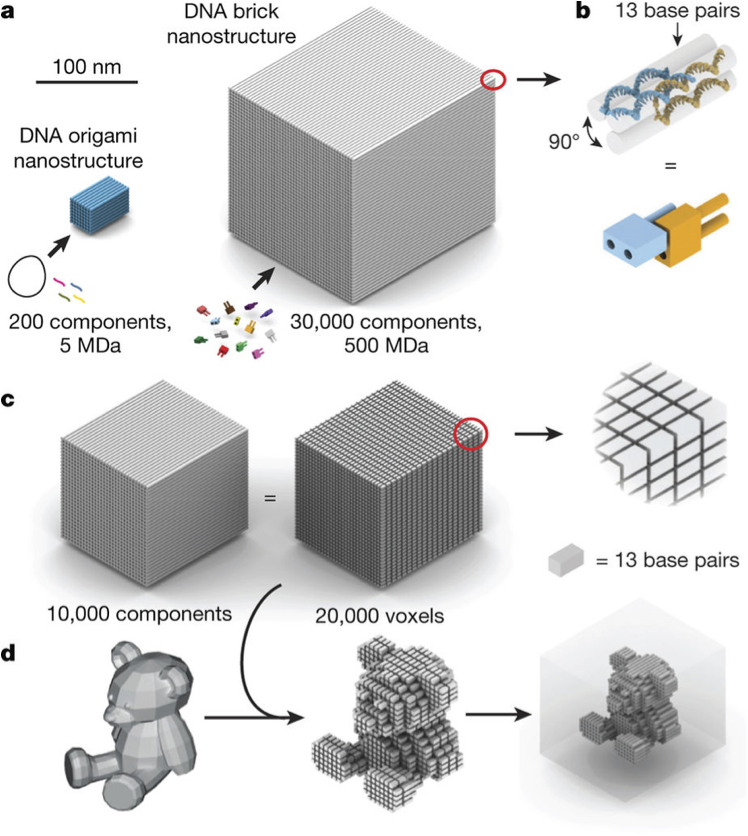
(a) 3D DNA origami are used to construct nanostructures with masses of around 5 MDa from about 200 unique components [circular scaffold (black) and staple (colored) strands>
. The DNA brick nanostructures assembled here have masses of up to 500 MDa and contain up to about 30,000 unique components (i.e. bricks). (b) Detailed helical (top) and brick (bottom) models of two 52-nucleotide DNA bricks bound to each other with a 90° dihedral angle via a 13-base-pair interaction. (c) A ~150-MDa DNA brick cuboid (left) consisting of about 10,000 unique components can be used as a molecular “canvas” (middle) with about 20,000 voxels (right), each containing 13 base pairs (see inset). The scale bar for a and c (shown in a) is 100 nm. (d) A 3D rendering of a 0.5 GDa teddy bear (left) can be approximated using the 20,000-voxel canvas (middle) to form the cavity of a cuboid structure (right).
Here’s what Ong et al. stated at the end of their publication:
“Even large DNA brick assemblies might be possible; the high cost of purchasing a large number of synthetic DNA strands restricted our testing to about 30,000 distinct bricks, but low-cost methods for synthesizing DNA strands (such as chip-synthesized DNA followed by parallel enzymatic amplification are available.”
One can only speculate—or dream—about what could be achieved in material science and technology with more massive 3D DNA assemblies constructed from larger DNA bricks. Whatever those achievements might be, I’m pretty sure they’ll be amazing, and won’t necessarily need to be conjured up by sipping a beer like Ned did for his “epiphany”.
As usual, your comments are welcomed.







