- This First of Its Kind Achievement Was Enabled by Nanopore Sequencing
- The Ski Trek to Icelandic Polar Hot Springs Took 11 Days of Sled Pulling
- Many Unknown Microbes Were Discovered
The title of this blog comes from a September 2019 publication by Gowers et al. In reading the study, I came to appreciate the scientific importance of being able to sequence fresh microbial samples at their source in the environment, rather than after freeze-storage and shipping to a distant lab. Before outlining this amazing technical achievement, I will discuss environmental microbial communities and sequencing at the source.

Why Sequence Microbial Communities on Site?
According to Escalas et al. in a 2019 review in Ecology and Evolution, microbial communities play key roles in nearly every biogeochemical process that makes Earth inhabitable. They mediate vital ecosystem processes such as primary production, decomposition, nutrient cycling, climate regulation, carbon storage, disease propagation, and pollutant transformation. In addition, microbes inhabiting the body of multicellular organisms are essential for the well‐being and survival of their hosts. Microbes interact and maintain dynamic relationships among themselves, formally akin to human communities, and the yearly number of publications about microbial communities keeps increasing, as seen in this chart.
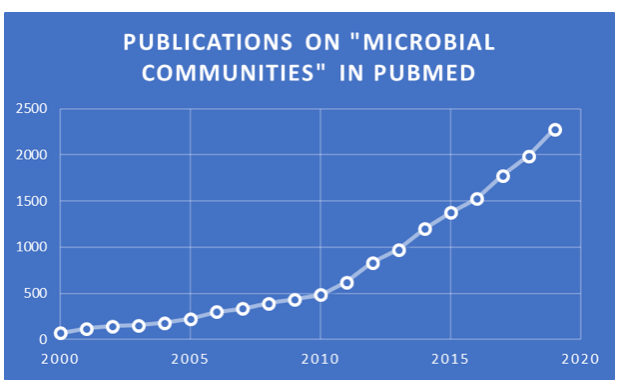 PubMed search results and chart by Jerry Zon
PubMed search results and chart by Jerry Zon
 Knowledge about this interactive and dynamic relationship among environmental microbial communities is correlated with accessibility for sampling, and the ability to obtain an unbiased representation of the resident microbes. Consequently, geographically remote or isolated areas are either not studied at all or to only a very limited extent. In particular, polar environments suffer from logistically challenging sample collection and shipping. According to a review, polar environments remain poorly understood at the microbial level, despite the fact that the polar cryosphere represents approximately 14% of the Earth’s surface,
Knowledge about this interactive and dynamic relationship among environmental microbial communities is correlated with accessibility for sampling, and the ability to obtain an unbiased representation of the resident microbes. Consequently, geographically remote or isolated areas are either not studied at all or to only a very limited extent. In particular, polar environments suffer from logistically challenging sample collection and shipping. According to a review, polar environments remain poorly understood at the microbial level, despite the fact that the polar cryosphere represents approximately 14% of the Earth’s surface,
Traditionally, microbial samples must be collected on site using sterile equipment and transported back to a laboratory under appropriate conditions, which frequently entail the samples being frozen under dry ice at -78 οC. Further logistic hurdles are encountered in clearing customs and other delays in transit. This often results in a significant length of time from sample collection to sample analysis, in which the composition of the microbial community can change or degrade.

Conventional characterization of microbial communities relies on DNA sequencing, since most microbes are unculturable. High-throughput metagenomic sequencing and amplicon sequencing are typically applied with the intention of revealing the proportions of microbial species present in a particular sample. Over the last decade or so, short-read sequencing has been the predominant method. However, this requires a full genomics laboratory to house large, bulky equipment. In recent years, long-read sequencing has seen increased use, but only Oxford Nanopore sequencing (depicted here) can produce significant amounts of long-read data (Mbp—Gbp) from a remarkably small device (MinION) capable of being run directly from a laptop, as pictured here.
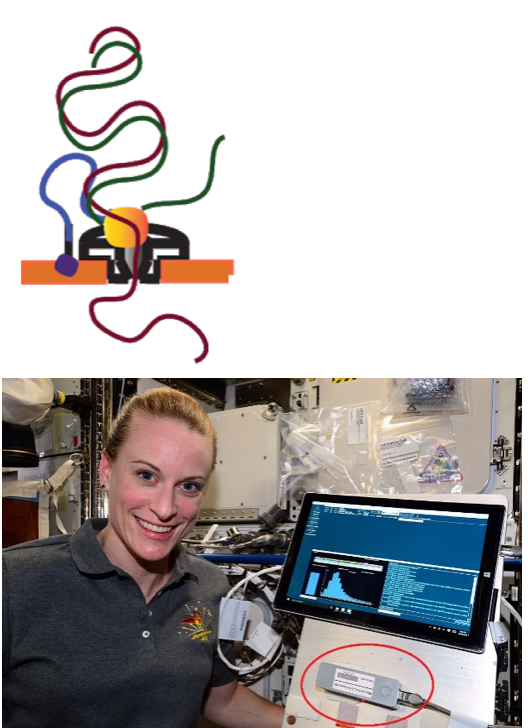
Taken from Workman et al. BioRxiv 2018. The copyright holder for this preprint is the author/funder, who has granted BioRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license and free to use.
DNA was sequenced in space as part of the Biomolecule Sequencer investigation on the International Space Station. The MinION sequencing device (circled in red) and sequencing was performed by ISS astronaut Kathleen (Kate) Rubins. Taken from commons.wikimedia.org and free to use.
Previous studies have capitalized on this device’s small size and portability, sequencing plant virus genomes in real time to inform crop management in sub-Saharan Africa, and to provide early detection and epidemiologic surveillance of Zika virus in Brazil. Minimalistic sample preparation is another factor that allows use of MinION in low-resource environments. Gowers et al. reasoned that “it is a clear progression to then apply these minimal-infrastructure techniques to polar environments,” [however>
“temperature regulation is a key challenge in
these environments as nanopore flow cells are extremely sensitive to freezing.”

The Trek by Ski and Sequencing by Solar Power
Sample prep and sequencing data collection were conducted in a tent after the 3-person expedition team (pictured here) had traversed 100 km of Europe’s largest ice cap over 11 days using solar power alone. The study occurred at least 60 km from civilization and at least 135 km from the nearest research facility. Crucially, everything required to conduct this metagenomic study was transported in a sled, along with all equipment required for long-term polar survival.


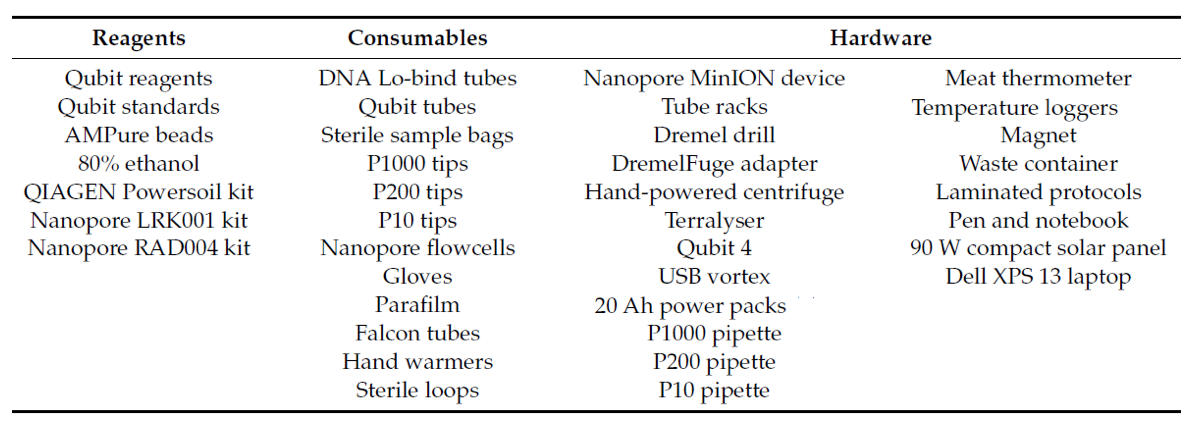
Detailed planning was required to ensure that absolutely everything needed for sample preparation and sequencing was carried or pulled on this expedition, since there was no way of obtaining anything that may have been overlooked. The entire portable laboratory setup was contained within two 9-liter boxes, in addition to a laptop (in a waterproof/shockproof case) and a 90 W solar panel, as detailed in this Table and shown laid out inside the tent.

During the ice cap traverse, three temperature zones were also established: kept on person, taken into the tent each night, and left outside in the sled. The nanopore flow cells were always kept on person since freezing destroys them. At night, they were kept in a sleeping bag, and during the day they were kept strapped to the carrier’s neck inside a travel bag contained in a generic airtight plastic food container with foam padding for thermal insulation. Temperature of the flow cells was constantly recorded using a battery powered automatic temperature logger. This methodology maintained a temperature between +9 οC and +30 οC despite ambient outdoor temperatures reaching -17 οC.

Samples for sequencing were obtained from the Hveragil hot spring gorge in Iceland, following a 60 km traverse of the Vatnajökull ice cap by ski and sled. A metal spoon sterilized with ethanol was used to collect the sample into sterile sample bags. The sample consisted of soil approximately 3 cm under the surface of mosses and liverworts, located on an overhanging rock 30 cm above the surface of the hot spring water (+60 οC). Samples were then transported on foot and ski at ambient environmental temperature to the tent sequencing lab located 3 km from the sample collection site, as pictured here.
DNA was extracted using a commercially available soil kit and centrifugation steps that were performed using a battery-powered drill (very clever!) equipped with a 3D-printed tube holder (pictured at this link). DNA was eluted using a sterile elution buffer, which was pre-heated to body temperature by being held in a gloved hand (simple but effective!). Extracted DNA was quantified using a battery operated commercially available fluorimeter. During sample preparation, hand warmers inside a closed sleeping bag were used to keep all solutions at about +20 οC.

Results
Within 5 h of sampling, 6.54 µg DNA was extracted into 100 µL by pooling and concentrating four separate extractions. As detailed in the publication by Gowers et al., various battery/power banks and other operational conditions were investigated to assess the length of time during which a flow cell would provide sequencing reads.
To determine the maximum amount of data that a single flow cell could yield in a single solar-powered run, an initial aliquot of the DNA library was added to a flow-cell, followed by two further additions of this DNA library to maximize cumulative throughput. Three battery packs and the laptop were charged the day before using the solar panel. Local base calling was disabled and screen brightness reduced to minimize power consumption. During daylight hours, power was fed into the laptop using the 90W solar panel. This second run yielded 113,699 reads and lasted 19 h 45 min.
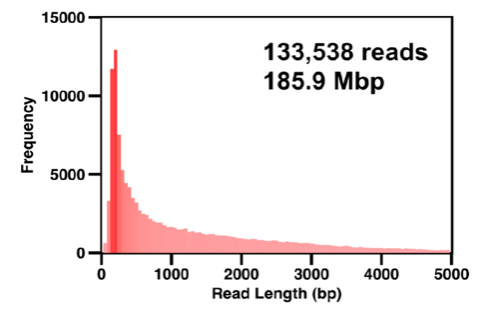
The total sequencing time for the expedition was about 24 h and yielded 133,538 reads and 185.9 Mbp of data. As shown here, a large number of “short” reads (dark red) resulted from aggressive bead-beating, which was required in the extraction of DNA from complex soil matrices, and the transposase step in the DNA library preparation. Nevertheless, a large number of reads extending to 5 kb in size and beyond were obtained.
All 133,538 base-called reads were pooled and uploaded for taxonomic classification. A total of 44% of the total reads (58,520) were successfully aligned to the reference database. This number is lower than was anticipated based on previous work, where values have fallen between 60% and 95%. Considering the remoteness of the sampling location, this likely represents uncharacterized microorganism species. A wide range of classes of bacteria were observed, as well as 2% eukaryotes and 0.4% archaea, as detailed in this chart.
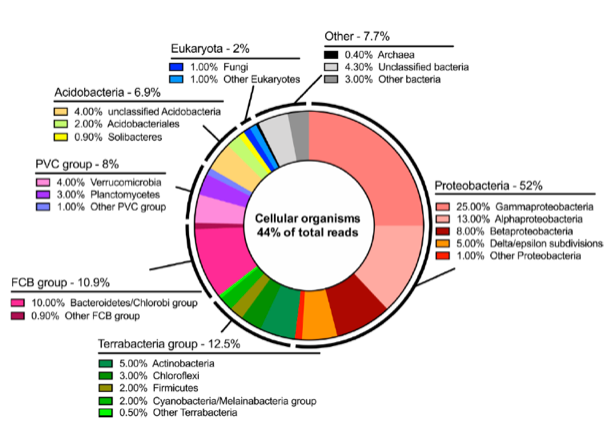
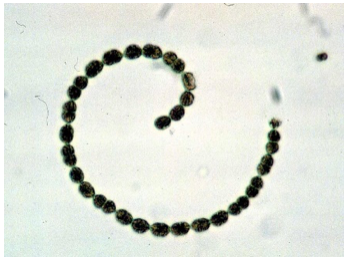
Due to sample collection at the edge of a geothermal stream, 10 thermophilic bacteria were identified by taxon classification of three or more reads (Chloracidobacterium thermophilum, Anaerolinea thermophila, Crenotalea thermop, Sphaerobacter thermophilus, Fontimonas thermophila, Spirochaeta thermophila, Symbiobacterium thermophilum, Schleiferia thermophila, Novibacillus thermophilus, and Pseudonocardia thermophila). Simultaneously, 1009 reads were assigned to organisms from the Cyanobacteria phylum with the Nostocales order (a member of which is pictured here) as the most abundant (371 reads). Within Cyanobacteria morphospecies, Nostoc (83 reads), Oscillatoria (29 reads), Phormidium (21 reads), and Pseudanabaena (9 reads) were identified. These are characteristic of microbial mats in polar regions.
Conclusions
“The dichotomy of microorganisms identified at Hveragil hot spring gorge was particularly intriguing,” according to Gowers et al., as “[t>
here was a sizable number of reads assigned to both cyanobacteria characteristics of the cold polar regions and thermophilic bacteria. The microenvironment present in hot spring streams fed by meltwater from the Vatnajökull ice cap is fascinating, as it gives rise to a microbial population with presumably extremely diverse metabolisms.
Gowers et al. add that “it is worth noting that approximately 56% of the data generated could not be aligned to the ‘NCBI nr + euk’ database. This number drops to 48% when a stringent quality threshold is applied to the reads. Therefore, most of this number likely represents organisms that have yet to be cultured and have their genomes characterized. While it may be expected that some of the data comes from unknown organisms, the numbers were surprising. Naturally, Hveragil gorge, on the northern edge of the ice cap, is among the most inaccessible places in Iceland. This, in combination with the interesting geochemistry of the area, leads to the conclusion that 56% represents a pool of interesting organisms and metabolisms that could be later isolated and studied further.”
Finally, they state that “the use of solar alone to power this sequencing effort demonstrated that microbial DNA sequencing can be conducted sustainably and repeatedly during an ongoing expedition, alongside the other physical and psychological challenges associated with survival in extreme environments. The method demonstrated here removes the need for sample transportation, and it is anticipated that this will enable researchers to study microbial populations in more remote corners of the planet, far from research stations.”
As I conclude this blog, I wish to thank Dr. Gowers for providing several pictures shown here, and for allowing access to a link to the teams’ website, which provides additional pictures and other information.
Your comments are welcomed, as usual.