An Interview with Dr. Shankar Balasubramanian about DNA G-Quadruplexes (GQs) Quantitatively Visualized in Human Cells for the First Time
My previous post featured Watson & Crick’s revolutionary publication of DNA’s double-helix structure in Nature in April 1953 resulting from their work in Cambridge. Now I’m fast-forwarding 60 years to January 2013 to highlight yet another landmark publication on DNA structure, again in Nature (Chemistry) and again based on work in Cambridge, but this time from a team led by Prof. Shankar Balasubramanian, who is The Herchel Smith Professor of Medicinal Chemistry at Trinity College, University of Cambridge.
The significance of Prof. Balasubramanian’s publication entitled “Quantitative visualization of DNA G-quadruplexes in human cells” coauthored with Giulia Biffi, David Tannahill and John McCafferty is evidenced by the fact that there were over 82,000 page views within 3 weeks of posting, as well as interviews with several news agencies, including the BBC! I’ve been very interested in GQs for quite some time so I reached out to Prof. Balasubramanian and he kindly agreed to my request for a telephone interview, which took place in March of this year. I’ve included some of my favorite questions from that interview below. For those of you who may be new to this area of research, I’ve included some background information on GQs as well as a postscript following the interview questions.
JZ: What inspired or led you to begin investigating cellular G-quadruplexes?
SB: My lab was actually working on the mechanism of human telomerase in the late 1990s and noted some interesting papers postulating the potential importance of G-quadruplex-folded (telomere) structure and telomerase function. This set me thinking about whether such structure might be ‘real’ in biological systems and how we might go about investigating them.
JZ: What has been the most challenging experimental aspect of this work?
SB: It is a culmination of much work done using molecular design and synthetic chemistry, antibody engineering, molecular biophysics and cell/molecular biology. The biggest challenge was to figure out how to usefully integrate and harness all of these sub-disciplines in the project.
JZ: Was there a “most memorable moment” for you during this research?
SB: Seeing is believing. Observing the first images of G-quadruplex hotspots in cellular genomic DNA was an exciting moment.
JZ: What was a “big surprise” during this research?
SB: Seeing the levels of G-quadruplex signal go up on addition of a G-quadruplex-stabilising small molecule. Whilst this was in accord with the hypothesis that G-quadruplexes can serve as targets, I hadn’t expected such a visually clean outcome from the experiment.
JZ: How will it be possible to target G-quadruplexes in specific DNA promoters?
SB: To target G-quadruplexes associated with particular genes one must consider the molecular structure of that G-quadruplex, which can be done biophysically, and additionally how the functional state of that part of the genome (e.g. is it transcriptionally active) makes it more vulnerable towards small molecule ligands. It is important to consider a G-quadruplex (or a cluster of G-quadruplexes) within the architectural context of the genome and chromatin.
JZ: Any guesses about the DNA G-quadruplex epitope for the B4G antibody?
SB: We don’t, as yet, have high-resolution structural information on a protein-DNA complex. Given the antibody recognizes a wide variety of G-quadruplexes we have looked at, it would appear that it recognizes common molecular features such as those of the stacked G-tetrad core, rather than the hypervariable loops.
JZ: Is the BG4 antibody available to other research groups now or in the future?
SB: The plasmid will be made available to others for research purposes via a standard MTA with our university.
JZ: Do you foresee possibly visualizing DNA G-quadruplexes in living cells in real-time?
SB: We and others are taking steps towards enabling this through the design and development of new molecular probes and I expect we will soon see how it can be done.
JZ: What is currently most controversial about DNA G-quadruplexes?
SB: I would say whether they have biological ‘function’. This will of course also depend on one’s strict definition of function and what constitutes evidence of biological function. I consider there is already good experimental support that there are biological consequences of G-quadruplex formation and/or stabilization.
JZ: You have a historical connection with inventing what is now Illumina’s sequencing-by-synthesis—have you applied this technology in your G-quadruplex research?
SB: Indeed we have. Last year we published a paper in which we used Solexa/Illumina sequencing to map the genomic sites of double strand breaks induced by a G-quadruplex stabilizing ligand. Gratifyingly these sites were mapped to regions richly populated by predicted G-quadruplexes. More recently, we have been mapping G-quadruplex structures using an antibody to perform DNA immunoprecipitation followed by deep sequencing.
JZ: Do you think it’s appropriate to refer to this field as “GQnomics”?
SB: I’m not one for expanding the ‘omics’ lexicon. However, I do think it is time for biologists and genome folk to consider how this non-Watson-Crick structural motif might play a role within their systems or questions under investigation.
Additional comments by Prof. Balasubramanian and photos are available in his interview with BBC News.
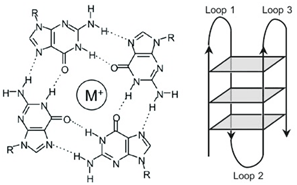
DNA guanosine (G)-quadruplexes (GQs) of the type shown below have been extensively studied from a chemistry perspective for many years. However, investigations aimed at convincingly demonstrating that DNA GQs are present and have functional roles in cells are experimentally challenging and not without controversy. Obtaining data and interpretation is difficult in part due to the large number of possible structural motifs that can exist and be involved in complex dynamic equilibria.
Figure 1: GQ Structural Formation
Prof. Balasubramanian’s publication on quantitative visualization of GQs in cells is the most recent contribution in a long line of his reports systematically elucidating the identity and function of this structural element. His early bioinformatics analysis of the human genome with Julian L. Huppert involved “putative quadruplex sequences” (PQS) defined and located by proposed working rules and search algorithms. They concluded that, in principle, as many as 370,000 PQS could exist simultaneously but that of these a smaller number would likely exist in a dynamic equilibrium with other structural forms of DNA. Importantly, it was found that PQS were present in a number of G-rich promoters throughout the genome and led to proposing that promoter GQs are directly involved in the regulation of gene expression.
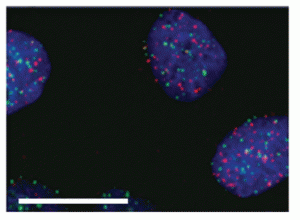
But can putative GQs be detected and studied in cells? That critical question was answered in the affirmative by Prof. Balasubramanian and his coworkers in their publication noted above. Quantitative visualization in fixed cells was achieved using immunofluorescence microscopy enabled by a DNA GQ-specific single-chain antibody (BG4) that they obtained from a phage display library of more than 2 × 1010 different antibody clones. Importantly, various BG4-enabled visualization experiments established that GQ structures are present largely outside telomeres (Figure 2). Moreover, quantitative visualization results obtained with synchronized cell demonstrated that GQ formation in DNA is modulated during cell-cycle progression—maximum BG4 foci per nucleus at S (synthesis) phase—and that these structures can be stabilized by a known GQ-binding small-molecule, pyridostatin (PDS). The authors state that it was “anticipated that G-quadruplex formation most probably occurs during DNA replication, because the associated mechanisms necessitate that duplex strands become separated at replication forks, where single-stranded DNA may fold more easily into secondary structures.” Stabilization of GQs by PDS was said to allow for possible use of stabilizing ligands to target cellular GQs and intervene with their function.
Figure 2: Immunofluorescence for BG4 showing absence of large co-localization between telomeric TRF2 proteins (green) and G-quadruplex (red) in U2OS (osteosarcoma) cells. This suggests that endogenous G-quadruplex structures are present largely outside telomeres. Nuclei are counter stained with DAPI (blue). Scale bar, 20 µm. |
Shifting molecular gears, so to speak, I encourage you to read a recent review coauthored by Prof. Balasubramanian on the occurrence and role of GQ structures in 5’ untranslated regions (5’-UTRs) of mRNA with regard to translational regulation and targeting. This emerging science is a fascinating and relatively new aspect of increasingly complex regulatory mechanisms involving RNA, some of which will be highlighted in a future post.
After researching the literature in this post, I have a far greater appreciation for the breadth and significance of molecular and cellular biology ascribed to GQ structures. So much so that at least for me there is a fascinating field of “GQnomics” in addition to genomics!
Comments about this post are welcomed!
This and future occasional Postscripts are a way to share information found when researching topics and technical details for my posts that I think are interesting or in some way noteworthy.
For example, I learned that identification of GQs had its origins in finding surprisingly stable aggregates of short synthetic poly(dG) oligonucleotides described in 1962 by a team led by H. G. Khorana, who incidentally 6 years later was awarded a Nobel Prize with Robert W. Holley and Marshall W. Nirenberg for their interpretation of the genetic code and its function in protein synthesis. Only a few months after the poly(dG)-aggregation report by Khorana and coworkers in 1962, Gellert et al. proposed that this phenomenon was due to planar, stacked arrays of four H-bonded Gs as shown above. Coincidentally, 1962 was also the year when Watson & Crick were awarded their Nobel Prize.
Who knew then that GQ structures would be found as telomeres that maintain chromosome stability in all living cells, and become a key part of the science for which a Nobel Prize in 2009 was awarded to Profs. Elizabeth H. Blackburn, Carol W. Greiner and Jack W. Szostak—a video interview of these Nobel Laureates is well worth visiting to hear how these scientists came to investigate telomeres and co-discover telomerase, which is a ribonucleoprotein enzyme that adds DNA sequence repeats ("TTAGGG" in all vertebrates) to the 3' end of DNA strands in the telomere regions. About 35 minutes into this 2009 video interview they are asked about potential therapeutic benefits of studying telomerase and telomeres. Profs. Blackburn and Greiner, who opine in response, co-founded Telome Health Inc. (THINC) one year later as a company that is “dedicated to leveraging the predictive power of telomere and telomerase assays (‘telome measures’) to assess health status, disease risk, and responses to specific therapies.”
One example of telomere-related therapy is a 13-mer oligonucleotide having a thio-phosphoramidate backbone named Imetelstat (GRN163L) that is in clinical trials by Geron Corp. Another is a phosphorothioate oligodeoxynucleotide called Cantide, which is antisense to human telomerase reverse transcriptase (hTERT; a catalytic subunit of telomerase) and has been recently reported to have promising anticancer activity in a nude mouse model of human primary hepatic lymphoma. Notwithstanding all of the above positive roles for GQs in biology and therapeutics, avoiding “runs of G” is well-known in design of PCR primers. This extends to antisense sequence selection, as exemplified by my investigations of a 15-mer having a run of 4 Gs that was seriously complicated by reversible intermolecular GQ formation evidenced by size-exclusion chromatography.
Comments about these Postscripts are also welcomed!