- Eric T. Kool Has a Long-String of Widely Cited Papers on Innovative Nucleic Acid Chemistry
- Kool’s Latest Contribution Involves Reversible Chemistry for PhotoCloaking RNA
- Kool’s Application of This to Aptamers is Very Cool, In My Opinion
 Eric T. Kool (taken from Stanford.edu)
Eric T. Kool (taken from Stanford.edu)
Readers of my blog know that my writing style tends toward using alliterations, which is evident in this blog’s title referring to Prof. Eric T. Kool and his chemistry, and that cool is the slang sense, i.e. awesome, swell, nifty, etc.
At the outset, I should say that Stanford University-based Kool has a long string of very ingenious publications in diverse aspects of nucleic acids, which attract many readers evidenced by ~1,400 citations per year according to Google Scholar metrics. Kool has received numerous awards and honors that are listed at The Kool Lab website, together with his lab’s research interests.
An appreciation of the diversity of Kool’s scientific contributions can be gleaned by later perusing his ~270 publications at this PubMed link, but for now I’ll be focusing on his most recent publication titled RNA Control by Photoreversible Acylation. This article requires—unfortunately—a paid subscription to J. Amer. Chem. Soc. or purchasing access to a pdf, so I’ve tried my best to give you herein a synopsis of what Kool has achieved in this study, and why it’s so cool.
Caged Nucleic Acids
One of the ways to externally initiate (i.e. turn on) nucleic acid biochemical or biological function in vitro or in vivo is to use a “caging” strategy. The term caging generally refers to installation of one or more removable groups on a molecule in such manner so as to render the molecule inactive. For nucleic acids (and proteins, etc.) this has typically been achieved with photoremovable groups, such that irradiation with UV light removes the caging group and restores functional activity.
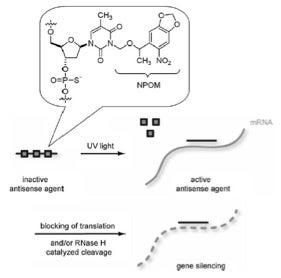 Taken from Deiters and coworkers ChemBioChem 2008
Taken from Deiters and coworkers ChemBioChem 2008
An illustrative example of this, published by Deiters and coworkers in 2008, is shown here for controlling gene silencing in mammalian cells with light-activated antisense agents. In this case, phosphorothioate-modified antisense agents were rendered inactive by installation of NPOM groups on nucleobases to prevent hybridization to a target mRNA. Brief irradiation with light at 365 nm removes the NPOM caging groups, enables sequence-specific binding to mRNA, and thus, blocks translation and/or leads to RNase H-catalyzed mRNA degradation.
A similar approach has been applied to functional studies of RNA. However, obtaining such photocaged DNA or RNA has usually required synthesis of chemically modified phosphoramidite monomer building blocks for solid-phase synthesis of the desired oligonucleotides. This limits utility to investigators who are able to deal with organic synthesis or have budgets to purchase relative costly photocaged monomers or custom synthesized photocaged oligonucleotides. Applying photocaging to long RNA is even more problematic.
Kool’s Cool Chemistry
To address these obstacles, Kool set out to develop a general method that can be applied post-synthetically to synthetic and native RNAs regardless of length, and moreover could be easily carried out by non-chemists. Based on studies (cited earlier) by others showing that 2’-OH groups of RNA can be selectively acylated in aqueous buffers with activated acyl compounds, Kool envisaged a strategy depicted here. An appropriately designed “PhotoCloaking” agent (PCA) is reacted with 2’-OH groups post-synthetically to block structure and interactions, but are rendered photo-responsive by including photocleavable bonds.
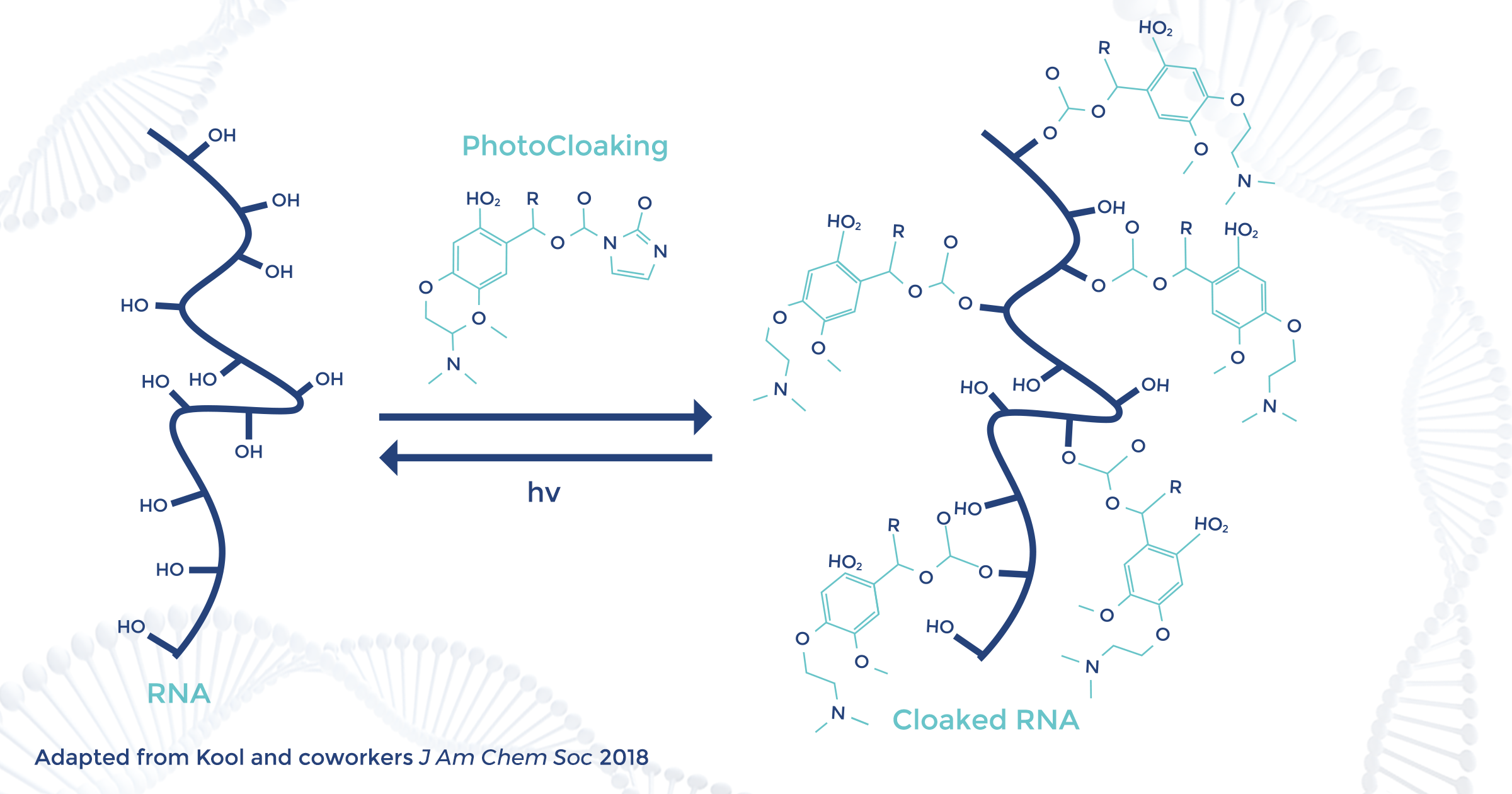
In his publication, Kool reasoned that “addition of several such blocking groups to the RNA (‘cloaking’) would in principle cover and protect it from folding and interacting with other molecules. Subsequently, the acylation could be reversed by exposing RNA to light, removing the blocking groups and switching on activity.” The PCA design strategy included combination of a reactive acyl group with an o-nitroveratryl photo-labile group, as shown here for PCAs 1 and 2 having unmethylated and methylated veratryl groups, respectively, to control the efficiency of photocleavage. Also included was a dimethylaminoethyl group to enhance solubility, and (after testing) 2-chloroimidazole as having the ideal level of reactivity.
A short 12-mer synthetic RNA oligo was used to evaluate 2’-OH acylation in water under mild conditions, in conjunction with denaturing polyacrylamide gel electrophoresis (PAGE) and mass spectrometry (MS), which indicated polyacylation with up to five groups (aka labels). Next, uncloaking of RNA by photoremoval of the PCA labels in water by irradiation with 365 nm light was confirmed by PAGE and MS. A 16-mer synthetic RNA oligo was similarly polyacylated and shown by native gel analysis and UV to have diminished ability to hybridize to complementary DNA. The reactivity of a synthetic hammerhead ribozyme previously reported was similarly disabled by PhotoCloaking and subsequently restored by exposure to UV light.
Kool then hypothesized that this postsynthetic polyacylation strategy would offer a convenient and simple method for preparing a photocontrolled aptamer of any length, which I have touted as a useful class of compounds in previous blogs. Kool’s test for this was especially interesting, in my opinion, because he used a 150-nt RNA transcript called Broccoli. This oddly named RNA is an aptamer that folds into a compact tertiary structure which binds DFHBI dye giving rise to a strong increase in fluorescence, and thus serves as an RNA mimic of well-known green fluorescent protein.
Kool had to use the more reactive PCA 2 to prepare a suitably cloaked version of Broccoli, which exhibited very dim fluorescence compared to uncloaked (i.e. untreated) Broccoli. When the cloaked aptamer was exposed to UV light and incubated with DFHBI, fluorescence was completely restored to the level of the untreated sample. The capability to use this PhotoCloaking strategy to switch on RNA function in cells was assessed by transfecting a 237-nt cloaked Broccoli-dimer RNA construct into a human (HeLa) cell line and then exposing these cells to light. As can be seen from the results shown here, the cloaking and uncloaking worked just as intended.

Kool concluded that “a great advantage of this postsynthetic labeling is that it allows for one-step photoprotection of long, biologically relevant RNAs that are difficult or impossible to synthesize via solid-state oligonucleotide synthesis. Since PhotoCloaking can be achieved without specialized equipment, the new method could potentially find widespread use for study of the biology of RNAs.”
Very cool indeed!
As usual, your comments are welcomed.






