- The US is Experiencing an Outbreak of C. auris Infections
- Drug-Resistant C. auris has a 30% Fatality Rate
- C. auris Genomic Sequencing and DNA Diagnostics are Available
Backstory
 3D illustration of C. auris
3D illustration of C. auris
The genesis of blog content for the Zone almost always arises from diligently scouting scientific publications, either directly or through databases. This blog is an exception, as it was prompted by the discovery of an alarming news article claiming that “the New Jersey Department of Health says there are over 140 confirmed cases of a deadly drug-resistant fungus in the state,” namely Candida auris. The article adds that there are more than 800 confirmed cases nationwide, and that the US Centers for Disease Control (CDC) and Prevention says that more than one in three patients with an invasive C. auris infection dies.
I searched the PubMed database, in an effort to learn more about the how and why of this serious issue, and determine whether I could blog about it. The decision was easy once I saw the data. The chart shown here clearly shows the sharply exponential rise in the number of publications on C. auris since 2009, when it was first discovered and so named as a novel yeast isolate from the external ear canal of an inpatient in a Japanese hospital.
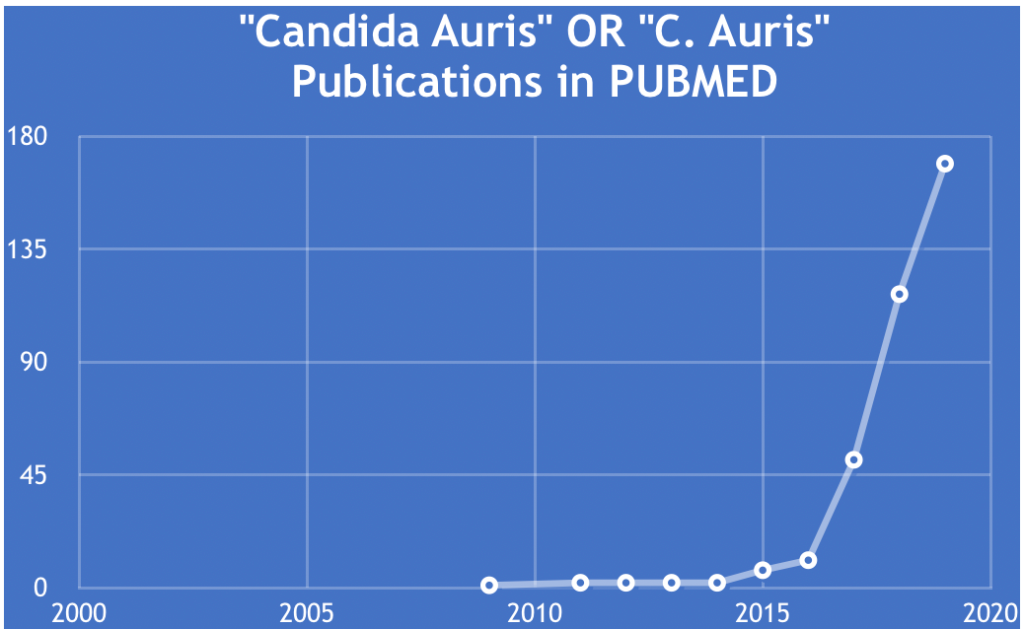 PubMed search and chart by Jerry Zon.
PubMed search and chart by Jerry Zon.
In keeping with the Zone’s focus on what’s trending in nucleic acid research, I will emphasize how C. auris DNA and RNA investigations have been used to elucidate the pathology and drug-resistant nature of this deadly fungus. But before getting into that, the next section will provide some noteworthy aspects regarding the epidemiology and origins of C. auris.
Epidemiology and Origins of C. auris
A 2018 review of the C. auris literature states that infection and colonization have been detected mainly in critical care patients, and affect both pediatric and adult populations. In addition, C. auris has been isolated globally, with information reported from South Korea, India, Pakistan, Kuwait, Israel, Oman, South Africa, Colombia, Venezuela, the US, Canada, and Europe, including the United Kingdom, Norway, Germany, and Spain. While reliable identification of C. auris is available in the US and other advanced countries, “issues with the identification of C. auris using both phenotypic and molecular techniques have raised concerns about detecting the true scale of the problem.”
 According to a 2019 publication by Jackson et al. at the CDC, “[t>
According to a 2019 publication by Jackson et al. at the CDC, “[t>
he reasons why this fungus began spreading widely in the last decade remain a mystery.” To address the mystery, they examined available data on C. auris and related species, including genomic epidemiology, phenotypic characteristics, and sites of detection, in order to put forth hypotheses regarding its possible origins. The following paragraph is a brief restatement of their reasoning, details of which can be accessed at this link.
C. auris has not been detected in the natural environment, although related species have been detected in plants, insects, and aquatic environments, as well as from human body sites. Importantly, it can tolerate hypersaline environments and higher temperatures than most Candida species. Jackson et al. offer hypotheses on the “pre-emergence niche” of C. auris both in the environmental microbiome and human microbiome, and speculate on factors that may have led to its spread. These include possible roles of healthcare, antifungal use, and environmental changes, including human activities that might have expanded the fungus’ presence in the environment or caused increased human contact.
For example, according to Jackson et al., it is plausible that farming of a particular crop in an area containing a C. auris ecological niche might lead to amplification, if the crop or its debris provided a suitable habitat for this fungus. Another possible contributing factor could be the use of fungicides, which reduce the fungal diversity in fields or in runoff, and therefore can select for resistant species like C. auris. Specifically, triazole plant protection fungicides (e.g. propiconazole), some of which share similar structural characteristics with triazole antifungal medications (e.g. fluconazole), have been increasingly used in crop agriculture in recent decades, and have been reported to induce cross-resistance to other Candida species.
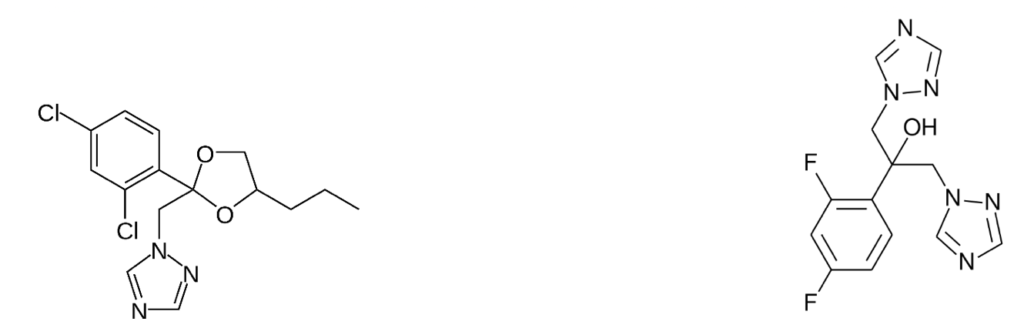
Genomic Insights on Multidrug-Resistant C. auris
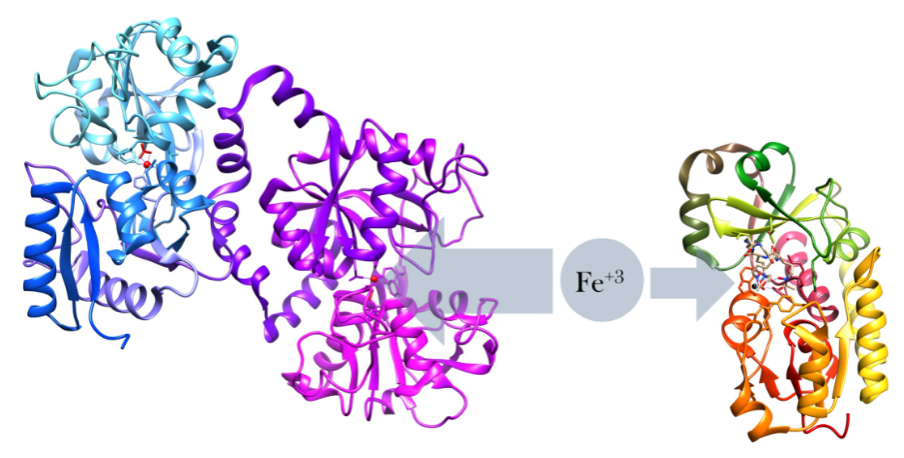
In 2017, Lockhart et al. reported that initial whole genome analysis of C. auris isolates from Pakistan, India, South Africa, Japan, and Venezuela identified four clades specific to each geographic region, suggesting that each of these clades emerged nearly simultaneously in different regions of the world. Muñoz et al. subsequently generated and annotated nearly complete genome assemblies for isolates from each of the four clades of C. auris, as well as for the three related species of Candida. Genomic comparisons revealed that C. auris has notable expansions of genes linked to drug resistance and virulence in C. albicans, including families of oligopeptide transporters, siderophore-based iron transporters (depicted here), and secreted lipases.
The larger protein, lactoferrin, is an immune-defense protein that prevents iron uptake from bacterial proteins, such as siderophores, the smaller protein. Lactoferrin does this by binding to iron with a higher binding affinity than most iron-binding proteins. Taken from commons.wikimedia.org and free to use.
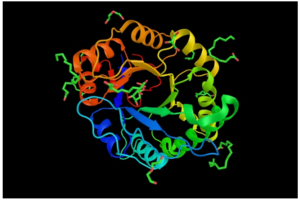
Muñoz et al. then used RNA-Seq to examine the response of two isolates of C. auris to antifungal drugs, as well as for the detection of the upregulation of transporters and metabolic regulators previously associated with drug resistance in C. albicans. They found that several either unique or expanded genes in C. auris and related species are upregulated > 2-fold (false discovery rate < 0.05) in response to two antifungal drugs, voriconazole and amphotericin B. Similarly, the expansion of lipases suggested that increased lipase production could be part of a shared mechanism of virulence, since it is known that the production of phospholipases (as exemplified here) helps in the adherence to and invasion of host cells, and is therefore relevant to Candida pathogenicity in the human host.
Molecular Diagnostics for Surveillance for C. auris
Thus, in order to implement appropriate infection control measures, both rapid tests for detecting patient colonization and broader detection capacities in clinical laboratories across the globe are needed, according to Kordalewska and Perlin. They argue that new molecular methods in C. auris surveillance methodology can substantially enhance detection capacity and facilitate outbreak response. In this regard, accurate identification of C. auris and differentiation from other yeast has been reported for Illumina sequencing of whole genomes. However, this method of sequencing may not be the first choice for routine determination of fungal species in some laboratories, mostly due to its technical demands, longer turnaround times, and substantial cost of instrumentation. The latter factor may be less of an issue for Oxford Nanopore sequencing of C. auris, as recently reported by Long et al.
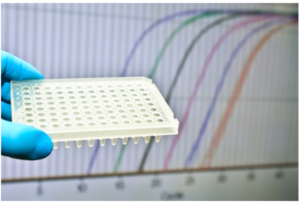
iven how ‘hot’ the C. auris topic is, it is not surprising to see that many research groups (academic and commercial) have developed a variety of molecular-based assays for the identification of C. auris.” Not surprisingly, PCR (end-point or real-time) is chief among these methods, ten of which are referenced. For example, Kordalewska et al. targeted ribosomal DNA (rDNA) regions using primer sequences designed to be specific for C. auris. A panel of 140 clinical fungal isolates was used for both end-point PCR and real-time PCR (depicted here), followed by electrophoresis or melting temperature analysis, respectively. The identification results from the assays were 100% concordant with DNA sequencing results.
Although not as popular as PCR, Yamamoto et al. have successfully devised and assessed the reliability of a loop-mediated isothermal amplification (LAMP) -based identification method specific to C. auris, enabling distinction of the pathogen from closely related species and other fungi. Their LAMP approach, which uses a device to measure turbidity (depicted here), distinguished all tested C. auris isolates (n = 20) from even very closely related species (n = 43) with a specificity of 100%. The assay was able to detect as low as 20 copies of target DNA per reaction. Readers interested in learning about the LAMP method can access an instructional YouTube video at this link.
The most revolutionary diagnostic technology that has been applied to C. auris is the T2 Magnetic Resonance (T2MR) system, which uses enzymatically amplified target DNA to sequence-specifically bind magnetic particles, as depicted elsewhere. This aggregation of particles leads to detectable modulation of the T2MR signal of water protons. Importantly, T2MR detection is culture-independent and can be performed directly 
Concluding Comments
Kordalewska and Perlin, who are experts in this area, summed up their opinions by stating that “[t>
he future of C. auris diagnostics is bright, with new molecular assays and technologies being developed each year.” These diagnostic advancements can substantially enhance detection capacity and facilitate outbreak response. With quicker and more efficient diagnostics, clinicians can make more informed treatment decisions. For example, clinicians could make data-driven decisions to limit the use of broad-spectrum antimicrobials, and thereby minimize chances for the development of resistance. Also, infection control specialists will be able to introduce more timely infection prevention and control actions.
Readers interested in learning more about infection intervention and control for C. auris in various settings, such as nursing homes, outpatient settings, and home healthcare, can consult a very informative CDC website at this link.
Your comments are welcomed, as usual.