- David Liu is Nature Magazine’s Number One Person Who Mattered in 2017
- Liu’s Team Has Modified CRISPR to Achieve Single-Base Editing of the Human Genome
- This Opens the Door for Developing Therapies for Tens of Thousands of Human Genetic Diseases
Nature magazine is considered by many—including yours truly—to be among the best sources of scientifically related news, publications, and editorials. Consequently, when I read the cover of a recent issue of Nature featuring its picks for the “ten people who mattered” in 2017, I was immediately intrigued, and felt compelled to read about who and why these persons were selected—especially for the uber-prestigious number one pick. You can read about all these folks later by clicking here, but for now I’ll focus only on David Liu, who was chosen by Nature as the number one person who mattered in 2017, which is a very special accolade.
The “Gene Corrector”
Referring to Liu as the “Gene Corrector” was Nature magazine’s way of concisely encapsulating the fact that Liu’s laboratory was able to take the already well-known CRISPR system, which I’ve blogged about extensively, to its highest possible pinnacle of performance. Namely, editing only a single nucleotide in the entire human genome comprised of six billion nucleotides, which is the ultimate in specificity for human genome editing. How this was achieved is briefly outlined in the next section, but before getting to that I think it’s nice to put a face with a name, and also include key contributors to this remarkable feat.
According to news from the Broad Institute, which along with Harvard and Howard Hughes Medical Institute are Liu’s multi-academic affiliations, this recent landmark achievement involved contributions by Nicole Gaudelli, currently a postdoctoral fellow in Liu’s lab; Alexis Komor, a former postdoctoral fellow in Liu’s lab who is now an assistant professor at the University of California San Diego; current graduate student Holly Rees; former graduate students Michael Packer and Ahmed Badran, and former postdoctoral fellow David Bryson.
 Holly Rees, David Liu, and Nicole Gaudelli (Credit: Casey Atkins). Taken from broadinstitute.org
Holly Rees, David Liu, and Nicole Gaudelli (Credit: Casey Atkins). Taken from broadinstitute.org
Evolution of Base Editing
Evolution of a “base editor” by David Liu has involved a progression of investigations that began in 2013 when Liu joined a host of other luminaries in founding a company now called Editas Medicine to develop treatments based on CRISPR technology. It became evident that despite the great potential for gene editing using CRISPR technology, clinical applications could be limited by the unpredictability of CRISPR/Cas9. Although the Cas9 enzyme cuts DNA where directed by guide RNA, researchers must rely on the cells’ own DNA-repair systems to fix the break, which can create a variety of different edits to the genome.
Liu’s lab looked for ways to improve on that. In 2016, then postdoc Alexis Komor and others on Liu’s team reported its first base editor. They used engineered fusions of CRISPR/dCas9 (a catalytically “dead” Cas9 mutant) and a naturally occurring cytidine deaminase enzyme that retain the ability to be programmed with a guide RNA. These fusion constructs do not induce dsDNA breaks, and mediate the direct conversion of cytidine to uridine, which is copied by polymerases into DNA as T, thereby thus converting a C•G base pair into a T•A within a window of approximately five nucleotides.
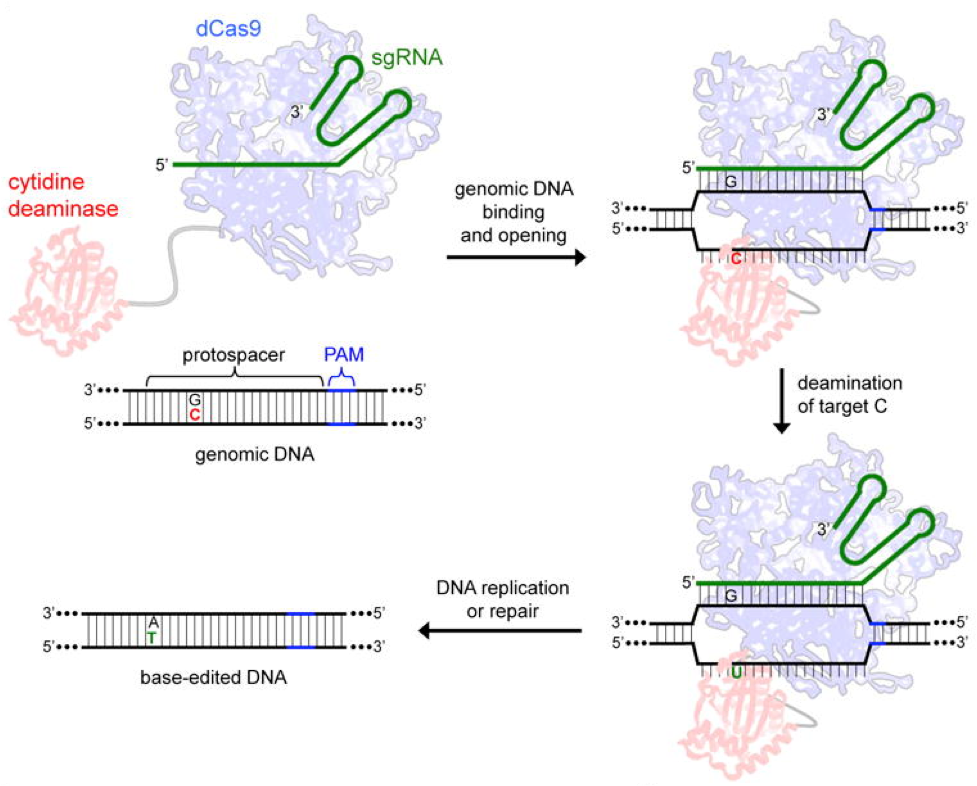 Taken from Komor et al. Nature 2016.
Taken from Komor et al. Nature 2016.
The approach has since been deployed in a range of organisms, from wheat to zebrafish and mice. And in September 2017, researchers in China reported that they had used Liu’s base editor to correct a single-letter mutation, or point mutation, for a blood disorder (β-thalassemia) in human embryos, although the edited embryos were not allowed to develop further.
According to the Nature article, Liu postdoc Nicole Gaudelli was eager to build on that work and create an analogous system that could instead deaminate adenine to produce inosine (I). Within the constraints of a polymerase active site, inosine pairs with C and therefore is read or replicated as G. The original A•T base pair is thus replaced with a G•C base pair at the target site. However, no naturally occurring enzymes are known to deaminate adenine in DNA. Nature adds that Gaudelli was therefore proposing to “break a cardinal rule in the Liu lab: no one takes on a project if the first step is to create a new enzyme. The risk of lost time and failure is too high.” Fortunately, Liu nevertheless encouraged her to pursue this risky challenge.
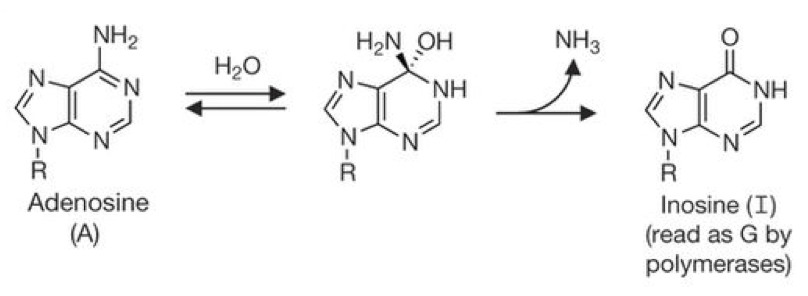 Taken from Gaudelli et al. Nature 2017.
Taken from Gaudelli et al. Nature 2017.
The Broad news article quotes Gaudelli as saying the main challenge for her while developing an adenine base editor (ABE) was “overcoming the psychological hurdle of whether or not ABE could go from concept to reality, since the key component of the editor did not exist naturally and had to be evolved in our lab. It was important to keep the faith that we could not only dream of such a molecular machine, but also build it.”
Building the required hypothetical deoxyadenosine deaminase turned out to be a difficult task that necessitated considerable, sophisticated molecular biological engineering that is more than I can succinctly summarize here. Interested readers will have to consult details in the publication by Guadelli et al. that describes the strategy and execution of numerous rounds of this enzyme evolution and engineering.
During this process they characterized the most promising ABEs from later rounds in depth by choosing a set of 17 human genomic targets that place a target A at position 5 or 7 of the protospacer (see above scheme) and collectively included all possible NAN sequence contexts, wherein N = A, G, C or T. The base editing efficiency of the most active editor (ABE7.10) overall averaged 53 ± 4% at the 17 sites tested, exceeded 50% at 11 of these sites, and ranged from 34–68%. These results were said to compare favorably to the typical C•G to T•A editing efficiency reported by Komor et al.
Guadelli et al. determined that the activity windows of late-stage variants are approximately 4–6 nucleotides wide, from approximately protospacer positions 4 to 7 for ABE7.10, and positions 4 to 9 for several other ABEs. Importantly, they concluded that “the precise editing window boundaries can vary in a target-dependent manner,” which to me implies that application of this methodology will require case-by-case genetic disease-specific optimization with potentially varying degrees of success.
Guadelli et al. did not detect any apparent ABE-induced A•T to G•C DNA editing outside on-target or off-target protospacers following ABE treatment. Although additional studies were said to be needed to examine possible untoward RNA editing by ABEs, they observed no elevated adenine mutation rate among four abundant mRNAs in ABE7.10-treated HEK293T cells compared to untreated cells, nor any apparent ABE toxicity in bacterial or human cells under the conditions investigated.
Editing Disease-Relevant Mutations
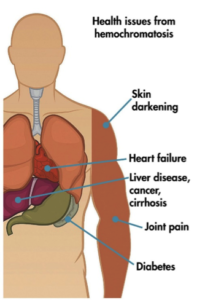
Finally, Guadelli et al. tested the potential of ABEs to introduce disease-suppressing mutations to correct pathogenic mutations in human cells. In one example, the genetic iron storage disorder hereditary hemochromatosis (HHC) was tested. HHC is commonly caused by a G to A mutation at nucleotide 845 in the human HFE gene, resulting in a C282Y substitution in the HFE protein that leads to excessive iron absorption and potentially life-threatening elevation of serum ferritin.
They transfected DNA encoding ABE7.10 and a guide RNA that places the target adenine at protospacer position 5 into an immortalized lymphoblastoid cell line harboring the HFE C282Y genomic mutation. Editing efficiency was measured by high-throughput DNA sequencing of genomic DNA, which showed clean conversion of the Tyr282 codon to Cys282 in 28% of sequencing reads from transfected cells, with no evidence of undesired editing or indels at the on-target locus.
Concluding Remarks
While the development of ABE is an exciting step forward in base editing, more work remains before base editing can be used to treat patients with genetic diseases, including tests of safety, efficacy, and side effects. “Creating a machine that makes the genetic change you need to treat a disease is an important step forward, but it’s only one part of what’s needed to treat a patient,” said Liu in the Broad news item. “We still have to deliver that machine, we have to test its safety, we have to assess its beneficial effects in animals and patients and weigh them against any side effects—we need to do many more things.”
“But having the machine is a good start.”
I fully agree, and hope that you will too.
As usual, your comments are welcomed.
Addendum
After writing this blog, it was announced that there is now a specialty journal dedicated to CRISPR-related investigations. The February 2018 inaugural issue of this new journal—aptly named The CRISPR Journal—is shown below.







