A Short Walk in the Wondrous, Wacky World of Long—and now Circular—Noncoding RNAs
I’m pleased by how much I learn when researching topics for new content, and this was certainly the case for long noncoding RNA (lncRNA), which was briefly mentioned in my last blog post, “Ripples from the 2013 TIDES Conference.” The topic piqued my interest so I set out to find out more. Plowing through lncRNA (aka lincRNA = large intergenic non-coding RNA) literature I quickly realized that there was an enormous amount of information, and the big challenge would be to capture some intriguing aspects without getting bogged down in “technical weeds” or being overly simplistic. In what follows there is a super brief introduction to what lncRNAs are and what they do—the latter is controversial—along with an appreciation for why lncRNAs are indeed a structurally and functionally wondrous class of nucleic acids that now encompass circular molecules. Maybe—to borrow from Forrest Gump—lncRNAs are like “a box of chocolates” for molecular biologists.
Introduction of lncRNA
The basic definition of lncRNA as “non-protein coding transcripts longer than 200 nucleotides” distinguishes this class of RNAs from various types of shorter RNAs, such as microRNA (miRNA) and well-known, Nobel Prize-related short interfering RNA (siRNA). But that’s where the simplicity ends and things get complicated. You might be thinking…hold on…why should I want to know more about lncRNAs? My reply to that is to consider the following points, each of which struck me as truly stunning.
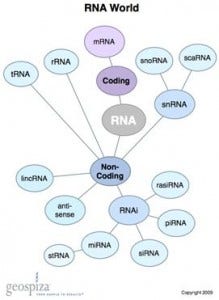
- Across the human genome there are four-times more lncRNAs than traditional coding RNAs. This large fraction of transcribed genome that extends beyond the boundaries of known genes is referred to as “pervasive transcription”—pervasive is defined in dictionaries as existing in or spreading through every part of something; why would cells do this?
- Whereas it was long held that only ~1% of human DNA is transcribed and therefore ~99% of the genome is “junk” DNA, new analytical methods have revealed quite the opposite: ~94% of the genome is transcribed, giving rise to a plethora of long (and short) ncRNAs.
- More than 1,500 lncRNAs are transcribed from two or more adjacent genes, and in some instances span chromosomal loci separated by more than 2 Mb! If you thought splicing of exons within a single gene is amazingly complex, then genesis of lncRNAs borders on bizarre.
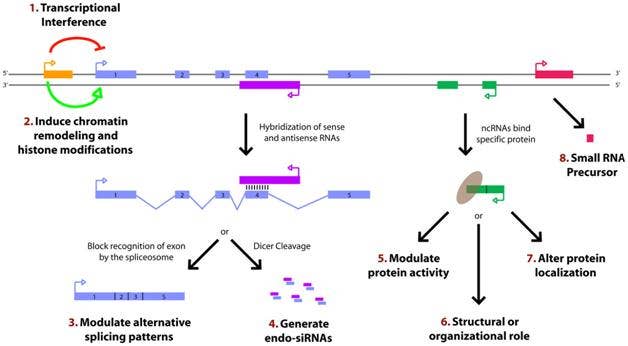 Paradigms for how lncRNAs function kindly provided for teaching by Cold Spring Harbor Press; for the explanatory caption click here.
Paradigms for how lncRNAs function kindly provided for teaching by Cold Spring Harbor Press; for the explanatory caption click here.
The Controversial Role of lncRNA
Lest you think that all is well in the proposed world of pervasive transcription, I hasten to add that this is definitely not the case. Bakel et al. have challenged the validity of tiling array data based on comparisons with data from RNA-Seq (aka whole transcriptome shotgun sequencing). They conclude that, “while there are bona fide new intergenic transcripts, their number and abundance is generally low in comparison to known exons, and the genome is not as pervasively transcribed as previously reported.” Clark et al. offer a lengthy and technically detailed rebuttal entitled The reality of pervasive transcription, to which Bakel et al. respond in lengthy technical detail.
Scientific debate is a healthy process, but non-experts face a difficult situation: how does one decide which of these opposing expert-views is correct? Doing more literature searching led me to a publication with a title—metaphorically linked to cosmologic “dark matter” — that directly addresses this issue, namely, Transcribed dark matter: meaning or myth by Ponting & Belgard. These authors propose that resolution of the debate requires “demonstration, or otherwise, that organismal or cellular phenotypes frequently result when noncoding RNA loci are disrupted.” In other words, if a lncRNA exists and has function, its knockdown by some method should ideally result in an effect that is somehow measurable. While this will obviously take much time and effort, Ponting & Belgard further opine that “[o>
nly then, when we have these data and the results of detailed mechanistic studies at hand, will dark matter transcription be revealed as either 'sound and fury, signifying nothing' [as it has been described>
or else as functional elements that are crucial to the biology of our species.” Who knew Shakespeare’s Macbeth would be thus quoted!
It has been remarked by Richard Robinson that “most dark matter transcripts are not signals emerging from a hidden universe within the genome, but instead simply the noise emitted by a busy [sequencing>
machine.” This view struck me as being extreme and possibly misleading, so I asked Piero Carninci, a leading expert in lncRNAs, for his opinion. He share’s it here:
“The rare non-coding transcripts are indeed truly transcribed, but in a much more tissue and cell specific manner, so they may appear as "background" transcription if the right cell is not selected. Therefore I do not think they are just ‘noise’ because of their specific expression pattern. Moreover, when looking at RNAs deriving by fractionating specific cell compartment (nuclei, chromatin, etc.) we see that these non-coding transcripts show very strong, specific compartment expression specificity. We are further characterizing the function of several of them and we see an important role in transcriptional control for several of them. Although we cannot claim that all of them are functional, we cannot state they are ‘noise’. Experiments will tell their ultimate functions.”
Readers interested in getting a sense of the extensive experimentation employed to unambiguously knockdown (or overexpress) a lncRNA and thus assess its biological function are directed to an investigation of a lncRNA called—would you believe—HOTAIR. This lncRNA reprograms chromatin and, in a brief review by Hung & Chang, is said to be overexpressed in ~25% of human breast cancers, drives metastasis in a mouse model, and is a prognostic marker for death and metastasis in human breast cancer. Association of HOTAIR and, moreover, a list of 166 other disease-related lncRNAs definitely makes for a “hot topic” (pun intended).
In wrapping up this blog, I wanted to add yet “more fuel to the fire” for ncRNAs that, until recently, have been linear molecules—long or short—by paraphrasing from an In Focus News article in Nature earlier this year entitled Circular RNAs throw genetics for a loop by Heidi Ledford. Basically, nonconventional sequencing of RNA led Memczak et al. to the discovery of thousands of well-expressed, stable, circular RNAs (circRNAs) that often show developmental-stage-specific expression. Tellingly, a ~1,500 (!) nucleotide-unit circRNA called CDR1as was found to have ~70 hybridization sites for a miRNA called miR-7 and, with other evidence, was shown to function as a molecular “sponge” that can sequester miR-7 and thus suppress activity of miR-7.
Ledford notes that circular RNAs have been hypothesized to also function by binding to viral microRNAs and RNA binding proteins, which suggests that circRNAs are a new class of regulatory RNA molecules. She quotes one researcher as saying that “[i>
t’s yet another terrific example of an important RNA that has flown under the radar.” Another researcher is said to hypothesize that “[t>
hey are so abundant, there are probably a multitude of functional roles.” Ledford concludes by asking Nobel Laureate Phillip A. Sharp what other shapes might RNAs take? To which Sharp responds: “I can’t think of another form we might have missed…[b>
ut you know somebody will find one.”
Hopefully, by now I’ve grabbed your attention and you’d like to find out more. Please check an lncRNA expert review by Piero Carninci and a continually updated database (lncRNAdb) containing a comprehensive list of lncRNAs that have been shown to have, or to be associated with, biological functions in eukaryotes. There is also a comprehensive analysis of human lncRNA gene structure, evolution and expression as of 2012 available in the GENCODE v7 catalog.
What do you think?
As always your comments are welcome.
Postscript
Henry Harris (University of Oxford) in the Correspondence section of Nature published online May 8, 2013 wrote the following in what was entitled History: Non-coding RNA foreseen 48 years ago.
The recent enthusiasm for studying non-coding RNAs (Nature 496, 127–129; 2013) brings to mind a largely forgotten review article that I wrote almost half a century ago in Evolving Genes and Proteins (V. Bryson and H. J. Vogel (eds) 469; Academic Press, 1965). This review reached a conclusion that was judged to be profoundly heretical at the time.
The article summarized years of work on the turnover of nuclear RNA, carried out during a period when pulse-labelled RNA was almost universally misdiagnosed as messenger RNA. It concluded: “Only a small proportion of the RNA made in the nucleus of animal and higher plant cells serves as a template for the synthesis of protein. This RNA is characterised by its ability to assume a form which protects it from intracellular degradation. Most of the nuclear RNA, however, is made on parts of the DNA which do not contain information for the synthesis of specific proteins. This RNA does not assume the configuration necessary for protection from degradation and is eliminated.”
Looking forward, not backwards, readers who wish to track future developments involving lncRNA can link to an lncRNA blog.
Also, there is a Keystone Symposium on February 27—Mar 4, 2014 called “Long Noncoding RNAs: Marching toward Mechanism” co-organized by Nobel Laureate Thomas R. Cech (1989; catalytic RNAs) and featuring a Keynote Address by Nobel Laureate Phillip A. Sharp, (1993; split genes, i.e. RNA splicing).