- The Virus is Quite Common with 267 Million Cases and 200,000 Deaths Annually
- RT-PCR is the Detection Method of Choice
- First Cell-Culture System May Speed Drug and Vaccine Development
We’ve all seen TV news stories about disgruntled passengers disembarking cruise ships returning to port early because of an outbreak of nasty gastroenteritis (i.e. inflammation of the stomach and intestines leading to nausea, vomiting, diarrhea, and stomach cramps). Norovirus (NoV) is the causative agent of these frequently reoccurring “nightmare” cruises, of which 13 have been reported since 2012, sickening some 200-600 passengers. It’s not just limited to cruises, the virus affected 100+ students at a school in Eugene, Oregon last year. And now there’s new evidence for transmission of NoV by eating oysters—which I will therefore not eat in the future.
 Taken from counselheal.com.
Taken from counselheal.com.
But perhaps the most NoV-related media attention—and investor ire or litigant action—has been recently focused on gastroenteritis outbreaks at Chipotle—a popular restaurant chain. A criminal investigation is under way at Chipotle, and according to an Associated Press report the company has been served with a federal subpoena.
Simple logic led me to me to think that if Chipotle has a problem with NoV, other big restaurant chains might also. And—more to the point—what’s the science behind NoV detection, treatment, and prevention? Following are some of that information I researched and found interesting, hoping you would, too.
Global & US Statistics
NoV reportedly affects around 267 million people and causes over 200,000 deaths each year. According to my math and the very scary—to me—Worldometers, these statistics translate to roughly 4% of the world population being sickened by NoV and 0.3% dying from it each year. These deaths are usually in less developed (the PC term for poor) countries, and typically affect the very young, elderly and immunosuppressed, who are unfortunately vulnerable to other health issues as well.
According to the Centers for Disease Control website—my “go to” source of reliable epidemiological facts in the US—each year on average in the United States, NoV is linked to:
- 19–21 million cases of acute gastroenteritis
- 1.7–1.9 million outpatient visits and 400,000 emergency department visits, primarily in young children
- 56,000–71,000 hospitalizations and 570-800 deaths
- 50% more norovirus illness in years when there is a new strain of the virus going around
Discovery
Being somewhat of a history-of-science buff, as well as an “ex-NIHer,” I was doubly pleased to find a first-person account by Albert Z. Kapikian describing how he and his colleagues in the Laboratory of Infectious Diseases at NIH first identified NoV in 1972. Interested readers should definitely consult this publication for details; however, a couple of “factoids” caught my attention.
First was that NoV got its genus name (i.e. noro) from the fact that samples from a 1968 outbreak in an elementary school in Norwalk, Ohio provided microbes ultimately identified as the causative agent. Second was the now seemingly primitive, but then state-of-the-art, methodology used to isolate this agent, namely rectal swabbing followed by filtration to remove bacteria (thus implying smaller, nanometer-sized particles typical of viruses). Since this was pre-PCR, and cultures were not possible, tracking the virus used volunteers who drank sample fractions and either developed symptoms or not—yuck!
Structure & RT-PCR Identification
Fast forwarding from those early discovery days to the modern era of molecular analyses gives us the luxury of now knowing that NoV are a genetically diverse group of single-stranded RNA, non-enveloped viruses that belong to five different genogroups—simply designated GI, GII, GIII, GIV, and GV—which can be further divided into different genetic clusters or genotypes. For example, GII, the most prevalent human genogroup, presently contains 19 genotypes.
Schematic representations of the relatively small—but nasty—7.6 kb genome and capsid assemblies are shown below. Further details can be obtained elsewhere. For this posting, however, it is noteworthy that this genetic diversity and ability to mutate significantly complicates detection and development of drugs or vaccines.
 Taken from viralzone.expasy.org.
Taken from viralzone.expasy.org.
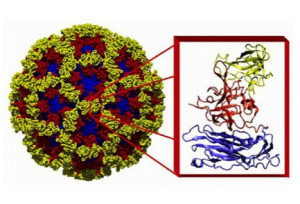 X-Ray structure of the NoV capsid, with the inset showing details of the structure of the protein subunits (RNA is inside, not shown). The different colors represent different regions of the capsid protein. Taken from bcm.edu.
X-Ray structure of the NoV capsid, with the inset showing details of the structure of the protein subunits (RNA is inside, not shown). The different colors represent different regions of the capsid protein. Taken from bcm.edu.
An extensive analysis of NoV mutation rates using hundreds of archived samples from around the world was published in 2013 by Kapikian and coworkers. Their research used RT-PCR, which involves transcription of NoV RNA into cDNA followed by PCR and detection (generally using kits similar to TriLink’s CleanAmp™ One-Step RT-PCR 2X Master Mix). They found that GII NoV maintained dominance over time, and also determined rates of nucleotide substitution/site/year for various strains, which is related to the interesting concept of “molecular clocks.”
My search of Google Scholar for NoV and RT-PCR in years 2014-present lead to nearly 2,000 (!) references, among which the following seemed interesting to me. I have hyperlinked these for convenience for those who also find the titles of interest:
- A single nucleotide polymorphism at the TaqMan® probe-binding site impedes the real-time RT-PCR detection of Norovirus 4 Sydney
- Fewer Multiplex real-time RT-PCR for the simultaneous detection and quantification of GI, GII and GIV noroviruses
- Detection of GI and GII noroviruses in drinking water and vegetables using filtration and real-time RT-PCR
- Survey of Canadian retail pork chops and pork livers for detection of hepatitis E virus, norovirus, and rotavirus using real time RT-PCR
- Inactivation of Human Norovirus by Common Domestic Laundry Procedures
Cell Culturing & NoV Drugs
As mentioned above, working with NoV has been severely hampered by lack of cell culturing and animal models. Fortunately, that is expected to change dramatically based on major new findings recently reported in venerable Science magazine.
In a nutshell, these investigators developed the first cell culture system for a human NoV (HuNoV) by finding that the current globally dominant GII.4-Sydney HuNoV strain infects human B cells—involved in immune response—that are accessed when NoV breaches intestinal epithelial cells. Surprisingly, this infection is substantially enhanced by certain carbohydrate-expressing bacteria present in the gut. It was concluded that “[i>
t is thus likely that previous attempts to culture HuNoVs failed because of the nature of the cell type tested and the absence of stimulatory carbohydrate molecules.” [Recall my blogs about how our lives are influenced by our gut microbiomes.>
Importantly, companion studies of related mouse NoVs validated that intestinal B cells are in vivo targets of NoVs, and that enteric bacteria are required for efficient infection of susceptible hosts. These collective findings set the stage, so to speak, for establishing defined mouse models and new drug-targeting strategies, such as blocking the aforementioned bacterial carbohydrate, as opposed to blocking NoVs RNA-dependent RNA polymerase (RdRp) shown in the above NoV genome. Regarding the latter approach, interested readers can peruse more than 50 references found by my Google Scholar search of NoV and nucleoside analogs from 2014-present.
NoV Vaccines
The familiar saying “an ounce of prevention is worth a pound of cure”—attributed to Benjamin Franklin—certainly applies to a vaccine to protect against NoV. Interested readers can consult a very comprehensive review and perspective I found by Kocher & Yuan published in 2015. Some snippets I found informative are as follows:
- Due to the inability to culture NoV, vaccine development has relied upon recombinant capsid proteins, such as virus-like particles (VLPs).
- Some VLPs have gone through clinical trials (e.g. LigoCyte acquired by Takeda Pharmaceuticals in 2012).
- An alternative strategy to produce VLPs utilizes viral vectors ( e.g. adenovirus). However, “biosafety concerns…may limit the development, availability and efficacy of the vector-based vaccine candidates.”
As I’ve commented on in a previous blog entitled Modified mRNA Mania, biosynthetic modified mRNA can now be investigated as a safe alternative to vectored vaccines. Let’s hope this is done soon, and is successful.
Your comments are welcomed.
Postscript

Another reoccurring food-safety problem worth noting here is listeria bacteria, which is recognized as one of the most virulent foodborne pathogens, with 20 to 30% of clinical infections (i.e. listeriosis) resulting in death. Responsible for an estimated 1,600 illnesses and 260 deaths in the U.S. annually, listeriosis is the third-leading cause of death among foodborne bacterial pathogens, with fatality rates exceeding even Salmonella and Clostridium botulinum.
Texas-based Blue Bell Creameries, which was forced to stop production and distribution of ice cream to 25 states back in April of 2015 due to listeria bacteria, recently reported that listeria was again discovered in one of their production facilities.

Readers interested in an illustrative example how PCR has been used to monitor and genotype listeria during food processing, as well as assess cleaning procedures, can consult a publication entitled Examination of Listeria monocytogenes in Seafood Processing Facilities and Smoked Salmon in the Republic of Ireland.






