- Two New Methods for Sequencing Pseudouridine Leverage Old Chemistry
- New Methods Reveal ‘Rewiring’ of Genetic Code by Post-Transcriptional Pseudouridination
- Exciting Future for New Analytical Methods for Modified mRNA
- Be Sure to Read the Very End of the Blog for a Special Offer!
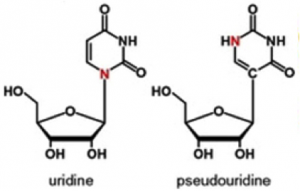
At the risk of seeming enamored with pseudouridine, which I previously proclaimed—with justifications—to be The 2014 Modified Nucleobase of the Year, recent reports about this fascinating base lead me now to feature it here once again. In that past post, it was pointed out that uridine, which is incorporated into all RNA during transcription of genomic DNA, differs from pseudouridine—historically abbreviated by the Greek symbol Ψ –by how one nitrogen (shown in red below) switches place with a carbon for bonding to the ribose ring. It was also noted that this switch has been long known to be carried out after transcription (aka post-transcriptionally) by an enzyme called—appropriately—pseudouridine synthase, the exact mechanistic details for which remain controversial. This post-transcriptional process that converts U to Ψ at specific positions in RNA is called pseudouridination.
 The bigger question attracting attention now is why “Mother Nature” evolved an enzymatic process for molecular manipulation of U to Ψ at only a few positions in various RNAs? To begin to decipher this puzzling evolutionary molecular biology, it’s necessary to have analytical methods to sort out where these positions are located, whether these loci change with cellular growth and differentiation, and the functional consequences thereof—definitely a daunting challenge.
The bigger question attracting attention now is why “Mother Nature” evolved an enzymatic process for molecular manipulation of U to Ψ at only a few positions in various RNAs? To begin to decipher this puzzling evolutionary molecular biology, it’s necessary to have analytical methods to sort out where these positions are located, whether these loci change with cellular growth and differentiation, and the functional consequences thereof—definitely a daunting challenge.
But, before plunging into recent reports on high-throughput, information-rich methods to pinpoint Ψs in RNA using modern sequencing technology, I’d like to briefly mention past work of others upon which these advances could not have been made.
Standing on the Shoulders of Giants
We should never forget that advances in science build upon work of others—a concept that goes back hundreds of years to a 12th century quote in Latin that translates to "discovering truth by building on previous discoveries"—as preeminent physicist Stephen Hawkins reminds us on the cover of his book about Einstein building upon Newton, Galileo, et al.
In researching the origins of site-specific location of Ψ in RNA, I came across many different contributors among whom James Ofengand gets my vote as providing—metaphorically—the strong, broad shoulders for later contributors to stand on. His many publications over four decades involving Ψ began in the Department of Biochemistry at the University of California San Francisco Medical School in 1962, followed by a 30+ year tenure at the Roche Institute of Molecular Biology in Nutley, NJ.
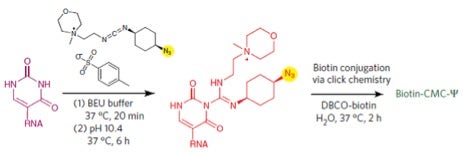
Ofengand’s 1993 report with Andrey Bakin in Biochemistry cleverly adapted Maxim-Gilbert sequencing, which relied on nucleobase-specific chemical cleavage of DNA, to work for RNA but used a different chemical for Ψ-specific modification. This chemical is the carbodiimide reagent shown below, which has a lengthy formal name that is thankfully abbreviated only as CMC. Skipping lots of chemically interesting—but far too “geeky” details—my simplified synopsis of their method is the following.
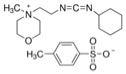 N-Cyclohexyl-N′- (2-morpholinoethyl) carbodiimide methyl-p-toluenesulfonate (CMC). Taken from sigmaaldrich.com.
N-Cyclohexyl-N′- (2-morpholinoethyl) carbodiimide methyl-p-toluenesulfonate (CMC). Taken from sigmaaldrich.com.
RNA is reacted with CMC under specified reaction conditions wherein U, G, and Ψ all form adducts; however, only the Ψ-CMC adduct will survive controlled alkaline hydrolysis of (i.e. removal of CMC from) U and G. The bulky CMC moiety now attached only to Ψs (at the N3 position) efficiently block reverse transcription of RNA into resulting cDNA. Therefore, 5’ radiolabeled primer extension leads to appearance of a gel band one base downstream from Ψ sites, and possibly at the site itself as a result of reverse transcriptase “stuttering,” while bands at U should be absent. A picture is said to be worth a thousand words, so I’ve provided one as part of the next section.
Back to the Future
Now, 20+ years after this clever CMC-mediated Ψ-mapping method by Bakin & Ofengand, an investigation co-lead by Thomas Carlile and Wendy Gilbert in the Department of Biology at MIT has gone “back to the future”—but not like the movie—to extend the original CMC method to contemporary massively-parallelized sequencing. The figure below taken from Carlile et al. in Nature 2014 depicts the sample preparation prior to sequencing using an Illumina HiSeq 2000 instrument in a method named—appropriately—Pseudo-seq.
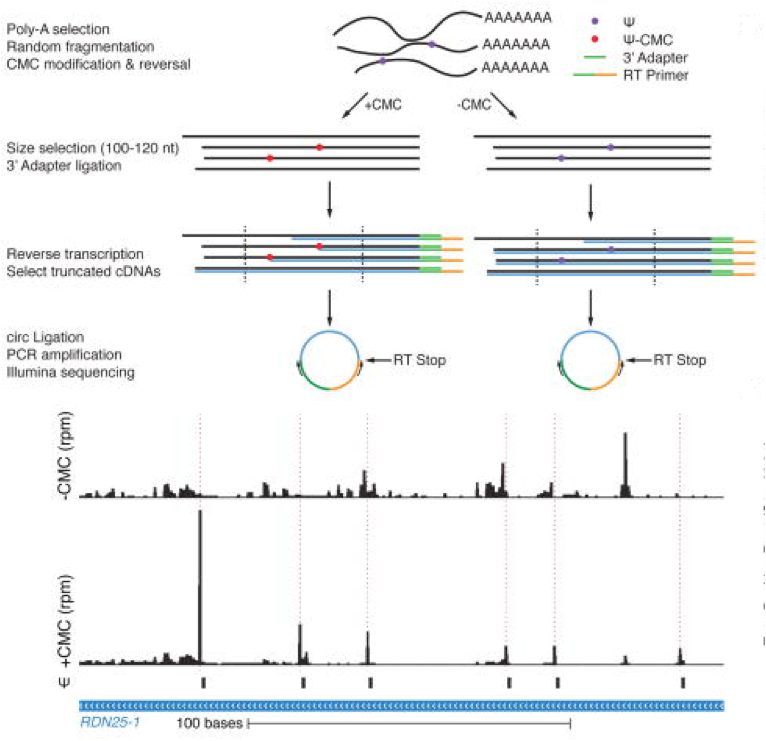
The method looks relatively straightforward; however, due to the relatively low abundance of of Ψs in RNA, Pseudo-seq requires quite a bit of work to reduce false-positive reads even with the use of +CMC and –CMC comparisons. For example, to define high-confidence Pseudo-seq reads in transcripts with sparse read-coverage, these investigators required reproducibility in 10-of-14 independent experiments! In this manner, for yeast, they identified 260 Ψs in 238 protein-coding transcripts.
Here’s the math I did to calculate average frequency of Ψ of mRNA based on these new data. In mammals the average length of mRNA transcripts is listed as 2,200-nt, therefore 260 Ψs in 238 mRNAS is ~0.04% frequency or 4Ψ-out-of-10,000-nt or 1Ψ-out-of-2,500-nt. This is on average only about 1Ψ per mRNA—but that can change; read on and learn why.
There are loads of other details in this Nature paper, but a couple of important take-home messages for me were the following. For the first time, it was shown that Ψ is present in yeast as well as human mRNA—previously, Ψ had been known to be present only in tRNA and rRNA. This finding adds Ψ to the short list of three modified nucleotides previously known for mRNA, namely, N6-methyladenosine (m6A), 5-methylcytosine (m5C), and inosine (I)—all available as 5’-triphosphates from TriLink. Importantly, the majority of Ψs in mRNA are regulated in response to cellular environment signals, such as nutrient deprivation in yeast and serum starvation in human cells. The authors state that "[t> these results suggest a mechanism for the rapid and regulated rewiring of the genetic code through inducible mRNA modifications.”
I’ve bolded this suggestion for emphasis given the remarkable implications of this “rewiring” mediated by yet another type of post-translational modification of mRNA that can modulate gene expression. I’m struck by how much more complex gene expression has been shown to be during the last decade or so, and can only wonder whether we collectively really do truly understand less the more we know! But I digress…
Variation on a Theme
As the PETA-insensitive but familiar saying goes, there’s more than one way to skin a cat. So, it’s not surprising that shortly after publication of the Pseudo-seq method, another team of investigators reported a “pull down” enrichment/sequencing variation—also leveraged on CMC, but with a twist; or better said with a click. Here’s why: they cleverly employed click chemistry, which I recently touted here—in the form of a new azido-CMC reagent that is “clickable” for biotin-mediated pull down prior to sequencing. Curiously—dare I say confusingly—this team of Chinese investigators named their method CeU-Seq as opposed to CeΨ-Seq.
New Sequencing Methods Needed for Modified mRNA
As previously commented on here in a post entitled Modified mRNA Mania, Ψ-modified biosynthetic mRNA, in conjunction with certain other modified nucleobases—primarily m5C—has been shown to have great promise as a new modality for mRNA-based therapeutics. Most of these investigations to date have used modified mRNA (aka mmRNA) wherein U has been completely substituted with Ψ, and similarly for other modified bases such as m5C. Recently, however, Hausburg et al. in Germany have prepared mmRNA with various ratios of Ψ and m5C to assess how such mmRNAs differ in terms of protein expression efficiencies and cell viability for human mesenchymal stem cells (hMSCs) and fibroblasts from different origins.
Those of you interested in the results of this study will need to read the paper. I’ll only comment on analysis of mmRNA transcribed using less than 100% substitution of the natural nucleobase, which in this study was approximately 50% (a near equimolar mixture of U, Ψ C, and m5C). Ideally, there would be analytical methods to determine the different sequence-motifs for these four nucleobases and the relative distribution since these can vary depending on transcription conditions and other variables associated with purification of mmRNA.
This seemingly impossible task is now partly in hand by virtue of Pseudo-seq to determine different U and Ψ motifs in mmRNA that can each be counted, in effect, to determine relative distribution. Similar analysis for m5C in mmRNA is possible, in principle, by using Pacific Biosystems single molecule real-time sequencing (SMRT). SMRT detection of m6A in a specific mRNA extracted from total cellular mRNA has been reported and results have been published using SMRT for m5C and other modified nucleobases in DNA. My scientific “crystal ball” also foresees extension of Oxford Nanopore Technologies approach to direct, electronic, single molecule RNA sequencing to include mmRNA.
While evolving these various sequencing methods to mmRNA still requires much effort, my past experience with therapeutic oligonucleotides indicates that this attention will eventually be required by corporate adherence to Good Manufacturing Practices (GMP), related FDA guidelines, and future need to analyze the fate of mmRNA in vivo.
Tell me why you’re a fan of Ψ (or any other nucleobase) and you may win a copy of Hawking’s book, On the Shoulders of Giants. I’ll randomly pick 5 people who leave comments below to win the book. Check back on October 12 to see if you’re a winner!