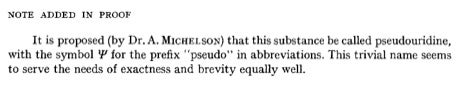- A Minor Modified Nucleobase Playing a Major Role in RNA Therapeutics
- Nucleobase is “Acrobatically” Formed in RNA by an Enzyme
- Its Original Identification is Linked to the Atomic Bomb!
The Molecule of the Year was started in 1989 by Science magazine and in 1996 was changed to ‘Breakthrough of the Year’. After a brief hiatus, the Molecule of the Year designation was revived in 2002 by the International Society for Molecular and Cell Biology and Biotechnology Protocols and Research. If you peruse the list of these lauded molecules, you’ll find that DNA has received this coveted award twice: “PCR and DNA polymerase” (1989) and “DNA repair enzyme” (1994).
While the Molecule of the Year is fascinating and well deserved, I think we should show some love for individual nucleobases given that this blog focuses on “what’s trending in nucleic acid research”—and since TriLink scientists are “The Modified Nucleic Acid Experts”. So, I thought it would be apropos (and fun) to start an annual post on Modified Nucleobase of the Year.
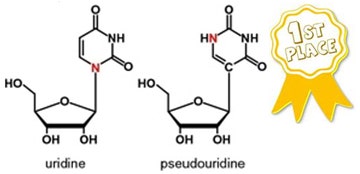
After mulling over which modified nucleobase merits this accolade for 2014, I decided on pseudouridine (aka 5-ribosyluracil)—an unusual isomer of uridine—for several reasons that are given below, followed by some history of its discovery that I found to be quite interesting.
Pseudouridine-Modified RNA Therapeutics
An earlier post entitled “Modified mRNA Mania” commented on relatively recent research demonstrating an exciting new paradigm in therapeutics based upon in vivo delivery of modified mRNAs to express target proteins. These proteins encoded by modified mRNAs have been applied to vaccines, cellular reprogramming, and other therapeutic modalities. In that post, I go on to indicate how such proof-of-concept publications have quickly spawned several new start-ups and mega-million dollar investments by Big Pharma companies. Although it’s still early days, the future for modified mRNA therapeutics is very promising.
Pseudouridine-5'-triphosphate (Pseudo-UTP) is used to impart desirable mRNA characteristics such as increased nuclease stability, increased translation or altered interaction of innate immune receptors. Pseudo-UTP, along with 5-methylcytidine-5'-triphosphate (5-methyl-CTP) has shown innate immune suppression in cell culture and in vivo, while actually enhancing translation.
For example, Warren et al. determined an efficient means of reprogramming multiple human cell types using modified mRNA that can express the four primary reprogramming proteins. These cells are referred to as induced pluripotent stem cells (iPSCs). Warren et al. found that modified mRNA substituted with Pseudo-UTP, 5-Methyl-CTP and ARCA effectively evaded the cell’s innate immune response, a crucial component in their success. Reduced toxicity due to substitution with Pseudo-UTP, 5-Methyl-CTP was critical since it allowed repeated transfection with modified mRNA over several weeks.
Readers interested in these approaches can access two posters here by TriLink scientists and collaborators.
“Acrobatic” Enzymatic Synthesis of Pseudouridine from Uridine
From the structures of uridine and pseudouridine shown above, you’ll notice that the red-colored nitrogen (N) and a double-bonded carbon in uridine have “switched places” in pseudouridine. Whether and how nature does this switch via one or more enzymes are intriguing questions that have been extensively investigated for many years.
The non-controversial answer to the first question about “whether” this switch occurs naturally is that nature has indeed evolved an enzyme for isomerizing uridine to pseudouridine that is named—appropriately—pseudouridine synthase. The second question about “how” is less clear, if not controversial, to some. Here’s why:
Pseudouridine synthase achieves this “acrobatic” positional switch on uridine that is already incorporated in RNA, as opposed to during biosynthesis of monomeric uridine prior to RNA synthesis. Since only a small number of uridines in RNA are converted to pseudouridine, understanding which ones are isomerized has been investigated. Readers interested in this can consult a recent report by Sibert & Patton entitled “Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements.”
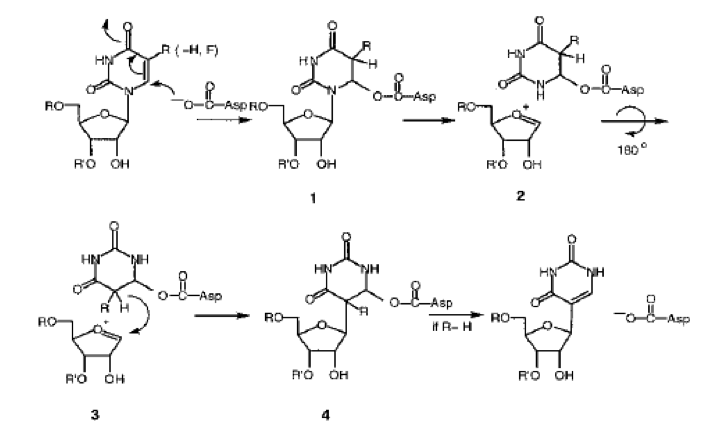
The controversial aspect of “how” deals not with sequence context but mechanistic details, which Santi and coworkers in 1999 proposed as follows, based on elegant substitution of hydrogen (H) with fluorine (F) at the C-5 ring position—BTW, my apologies for the organic chemistry “arrow pushing” but it’s a necessary evil to put up with to show the “how”.
On the other hand, subsequent work by Spedaliere et al. has led to questioning whether a different mechanism is operative. This is explored in detail in a publication entitled “The pseudouridine synthases: revisiting a mechanism that seemed settled.” Readers interested in this alternative view are encouraged to read that publication.
Pseudouridine-Modified Oligonucleotides
I hope this brief overview has spurred you to learn about the amazing things you can do with pseudouridine. Companies like TriLink offer custom chemical synthesis of pseudouridine-modified oligonucleotides that can be used for a wide variety of physicochemical, biochemical, or biological studies. Among biological applications, RNA interference (RNAi) using short, double-stranded RNA (aka siRNA) is especially important and widely employed. RNAi has been expertly reviewed by Beal & Burrows et al. with regard to incorporation of modified bases to probe and enhance RNAi.
Highlighted in the Beal & Burows review are comprehensive investigations by Nawrot and coworkers, who incorporated pseudouridine, as well as other modified nucleobases, at various positions to evaluate thermodynamic stability and gene silencing activity. As detailed in those results, certain locations for pseudouridine provided potent gene-silencing activity. I think you’ll see that these publications and reviews support my claim that pseudouridine is truly deserving of Modified Nucleobase of the Year status.
Fractionating Fractions and Chasing Spots
While I believe I’ve provided significant justification for my obesssion with pseudouridine, I understand that some of you may not be convinced. If the current range of potential uses and pivotal roles of this facscinating nucleobase haven’t been compelling enough, perhaps I’ll make you a convert when you learn more about this nucleobase’s origin.
Several groups of investigators in the 1950s were engaged in identifying minor constituents of RNA by developing then state-of-the-art chromatographic methods to separate hydrolyzed RNA into its components. Those of you who may not be familiar with column chromatography or paper chromatography simply need to know that eluted (aka separated) fractions are collected and further fractionated, or paper spots are cut out, followed by analyses to determine purity and identity. Sounds easy, but it’s not—especially in the 1950s.
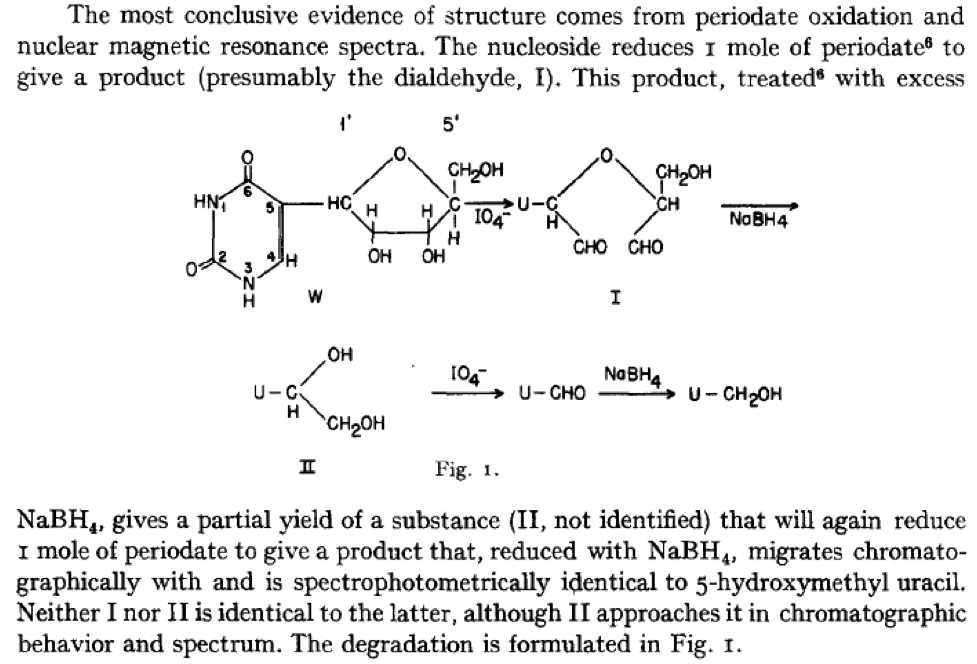
What these early investigators actually did for “fractionating fractions and chasing spots” of minor RNA constituents is way too complicated to get into here. So, I’ve taken the liberty to focus on the following work of Waldo E. Cohn, who has the distinction of being the first to definitively identify the very unusual structure of pseudouridine, and is associated with its naming.
Waldo E. Cohn: From the Atomic Bomb to Pseudouridine
In researching Cohn’s work leading up to identifying and naming pseudouridine, I learned about how his earlier involvement with the ultra-secret Manhattan Project to develop the first atomic bomb led to pioneering investigations of radioactive 32P biodistribution in rats! Here’s a brief excerpt on that taken from Simoni et al., which I’ll tie back to Cohn’s pseudouridine story.

By the mid-1930s, the cyclotron had been invented and developed at the University of California at Berkeley by Ernest O. Lawrence. With it came the capability of creating artificial radioactive isotopes. One of the first radioactive isotopes produced was 32P, which was immediately put to use in seeking solutions to biological problems.
Waldo E. Cohn was a graduate student at University of California Berkeley working with his thesis advisor Prof. David M. Greenberg. The 32P was prepared in the cyclotron of the Radiation Laboratory at Berkeley by bombardment of red phosphorus with deuterons. After purification and oxidation to H3PO4, the radioactive material was administered to rats, and the distribution of radioactivity in various tissues was followed with time, which you can read about here and is considered a “JBC Classic”.
Cohn's experiences with radioactive isotopes as a graduate student shaped his career. After receiving his Ph.D. degree in Biochemistry from Berkeley in 1938, and a postdoc at Harvard, he was recruited to the Metallurgical Laboratory of the University of Chicago in 1942 as a member of the Manhattan Project. His role was to study the biological effects of fission products on biological systems. Cohn moved to the Oak Ridge National Laboratory (ORNL) where he became Senior Chemist and Group Leader of the Oak Ridge Biology Division, a position that he held until his retirement in 1975.
Cohn was involved in the formation of the Isotope Distribution Committee, which was assigned the responsibility of making isotopes available to qualified researchers. In 1946, Cohn and Paul Aebersold published a landmark paper in Science that established the administration of isotope distribution and preparation of the catalog of isotopes that were available for research. It was one of the first post-war efforts toward the “peaceful use” of atomic energy. Cohn became well known for his efforts to establish and enforce standardized biochemical nomenclature. From 1965 to 1976, he was Director of the Office of Biochemical Nomenclature of the National Academy of Sciences.
Now back to pseudouridine!
The Right Scientist at the Right Place
Cohn’s experience with radioisotopes coupled with his expertise in nucleic acid-related chemistry and the unique resources offered by ORNL added up—in my opinion—to him being the right scientist at the right place to doggedly pursue identification of minor constituents of RNA.
Here are actual excerpts from his key publications on this hunt.
In concluding this post, I’d like to add one more historical “factoid” that I thought was interesting. I tracked down the identity of “Dr. A Michelson.” He is actually A. M. Michelson who coauthored with über-famous Sir Alexander Todd the first synthesis of ATP published in Nature in 1948. My thanks to Drs. Cohn and Michelson for their respective roles in giving “birth” to pseudouridine, which in turn has enabled development of new molecular tools for nucleic acid-based applications.
As we look at the historical significance as well as modern applications of pseudouridine, I hope I’ve convinced you of the amazing work this nucleobase is capable of. Let me know if you support my nomination for Modified Nucleobase of the Year, or share your nominations for other nucleobases below.