- Escherichia coli Bacteria are Used for Proof-of-Concept Studies
- Repetitive Centrifugation-Based Partitioning Enables Rapid Isolation
- Avidities of Found Aptamers are Comparable to SELEX Aptamers
Backstory
Aptamers are highly structured nucleic acids that bind to specific target molecules. DNA or RNA aptamers are usually selected in vitro from a very large pool (aka library) of random sequences, as discussed elsewhere. These pools of “randomers” are typically comprised of synthetic oligonucleotide sequences that have contiguous mixed-base (N = A/G/C/T or U) regions designated as Nn, in which n is typically 20-40 nucleotides. The mixed bases may also include chemical modifications to provide increased structural diversity, as exemplified by Pfeiffer et al. The random region is often flanked by fixed sequences for primer-based amplification and sequencing. Aptamer targets can be virtually any type of molecule, regardless of size, such as large proteins or small molecules, as shown here in the case of biotin, which is comprised of only 32 atoms!
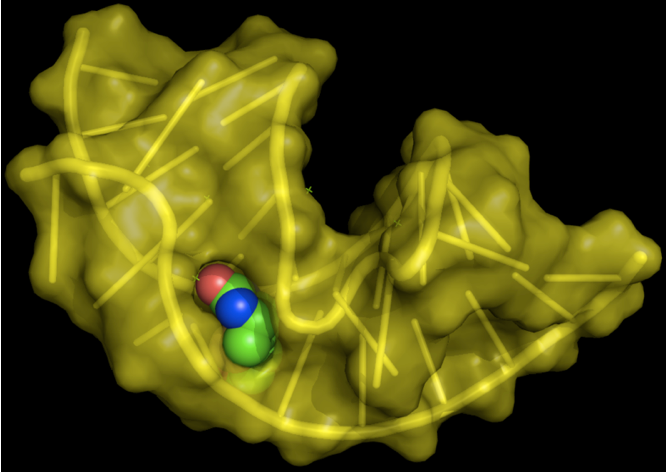 RNA aptamer (yellow) bound to biotin. Taken from commons.wikimedia.org and free to use.
RNA aptamer (yellow) bound to biotin. Taken from commons.wikimedia.org and free to use.
As a result of the ability to find aptamers for many sizes and types of molecular targets, the numbers of annual publications on aptamers and the myriad of applications they can be used for are increasing exponentially, as seen from this chart of data found in PubMed. This versatility has led to the widespread opinion that the utility of aptamers rivals—and sometimes surpasses—that of antibodies, as discussed in an earlier blog in the Zone. Indeed, aptamers are frequently referred to as “chemical antibodies” (e.g. Shigdar et al.).
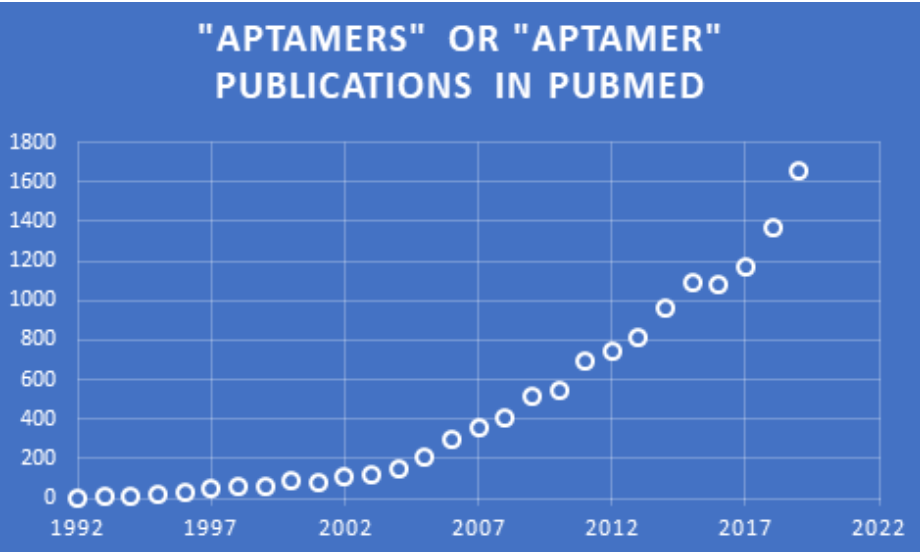 PubMed data found and charted by Jerry Zon.
PubMed data found and charted by Jerry Zon.
The vast majority of aptamers have been discovered by subjecting pools of randomers to a variety of selection methods, all of which stem from independent pioneering publications in 1990 by Ellington and Szostak in Nature, and Gold and Tuerk in Science. The latter investigators called their method “systematic evolution of ligands by exponential enrichment (SELEX),” now a broad descriptor in the aptamer field, as reviewed elsewhere. As of April 2020, this 1990 seminal SELEX publication has been quite remarkably cited over 9,100 times in Google Scholar.
Variants of SELEX applied to whole cells (aka Cell-SELEX) have become increasingly important as a means of finding aptamers that can function as cell-specific delivery agents or diagnostics. The first demonstration of Cell-SELEX was published in 1998 by Gold (see above) and collaborators, who used human red blood cells. An excellent and highly recommended review of the development of Cell-SELEX, including detailed protocols, was published by Tan and collaborators.
The next section provides further introductory comments on Cell-SELEX, followed by a synopsis of a recently reported variant that uses centrifugation as a simple separation step for selecting aptamers, as exemplified with intact bacteria.

Cell-SELEX
Aptamers for proteins were initially found using single purified proteins as targets. In contrast to this approach, Gold and collaborators demonstrated in 2003 that aptamer selection against complex targets, exemplified with a cancer cell line, is possible. They emphasized that in a Cell-SELEX approach, there is no need for prior knowledge of the molecular targets for the derived aptamers. As a corollary, they concluded that complex-target SELEX “[c>
an discover previously uncharacterized markers for tumors, pathologies, development, and differentiation. Such dissection of complex biological systems by the SELEX process may generate aptamers for diagnostic and therapeutic uses.”
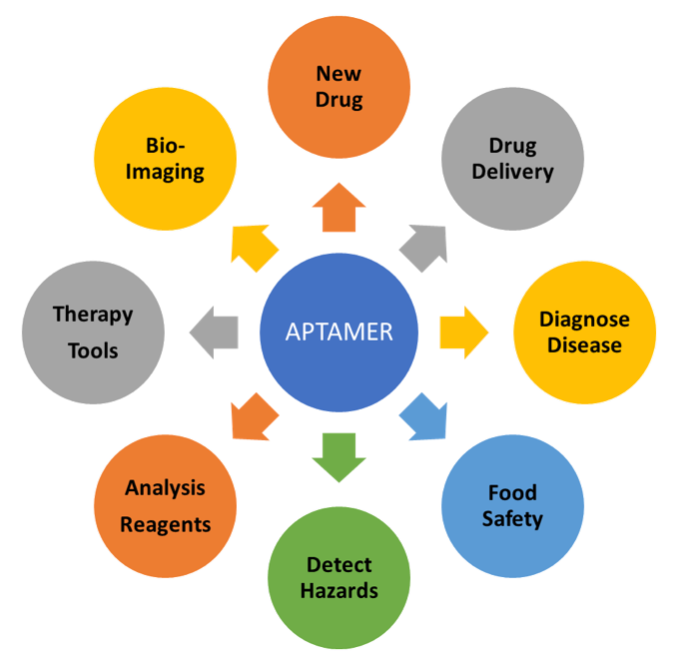
This prescient view of the potential power of Cell-SELEX has been realized, as reflected by nearly 240 articles in PubMed that are indexed to “Cell-SELEX” or “whole-cell SELEX” in all fields. Among these are over 20 reviews, which can be perused by readers interested in learning more about the scope of applications, including those depicted here.
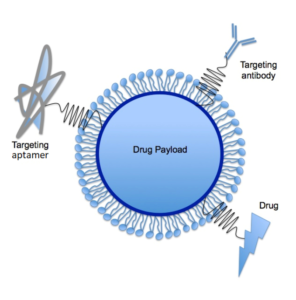
By way of example, Gourronc et al. have used Cell-SELEX to find RNA aptamers that internalize into human papillomavirus (shown here) type 16 (HPV-16)-associated oropharyngeal cancers, for which better therapeutic strategies are needed. The HPV-16 oncogenes E6 and E7 are expressed in HPV-associated cancers and are able to transform human tonsillar epithelial cells (HTECs). After 12 rounds of Cell-SELEX, a pool of aptamers with significantly greater internalization capacity (~5-fold) into E6/E7-HTECs compared to primary HTECs or fibroblasts was obtained. Aptamers that internalize into cells may be useful for delivering therapeutic agents to HPV-16 associated malignancies by, for example, guiding liposomes that carry a cytotoxic agent or an oligonucleotide/siRNA for oncogene knock-down by RNAi, as depicted here.
Centrifugal Partitioning

The idea for this blog arose from a pre-publication abstract of work by Dr. Byoung Chan Kim and coworkers in the Republic of Korea that described the first use of centrifugation partitioning as a faster and cheaper alternative to Cell-SELEX for selecting aptamers. These reported advantages seemed important enough to read in detail and feature in the Zone. An advance copy of the full article was requested from and kindly provided by Dr. Kim, pictured here.
According to Kim and coworkers, previously reported bacteria-specific aptamers obtained by Cell-SELEX usually include 7–30 cycles of incubation (i.e. binding), separation, and amplification of probes from randomer pools to enrich the oligonucleotides that are bound to the target cells, as shown in the process-flow chart here. Subsequently, enriched oligonucleotide pools from PCR amplification are cloned and sequenced. It was further stated that the repeated enrichment steps essential to obtaining high-affinity aptamers increase the cost and time associated with this process, and “require extremely large amounts of the target cells for aptamer isolation.”
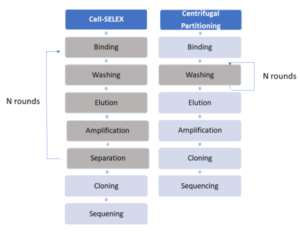
Unlike these methods, Kim and coworkers investigated a rapid way to isolate bacterial Escherichia coli cell-specific DNA aptamers, using centrifugation in a new manner, without requiring repeated rounds of elution and amplification of bound probes. This novel method is not called Cell-SELEX, and is instead named “repetitive centrifugation-based partitioning” between bacterial cells bound to DNA and unbound DNA. This centrifugal partitioning process-flow, shown above, enables the rejection of unbound DNA through sequential centrifugations of the initial mixture of target bacterial cells and randomer pools. The weakly bound DNAs are removed during the sequential partitioning. The DNA pools bound to cells are amplified only once prior to cloning.
Experimental Methods
Authentic samples of E. coli, Bacillus subtilis, Klebsiella pneumonia, Citrobacter freundii, Enterobacter aerogenes, and Staphylococcus epidermidis were obtained from the Korean Collection for Type Culture. The 88-mer oligonucleotides in the single-stranded randomer DNA pool were comprised of 45 random nucleotides (N), flanked by 19-mer and 24-mer primer-binding sequences at the 3′ and 5′ ends:
The randomer pool (1.8 nmol) was heated at 95 °C for 5 min, cooled on ice for 10 min, and mixed with 107 E. coli cells using a binding buffer and constant shaking (1000 rpm) at room temperature for 1 h. The mixture was then centrifuged (13,000 rpm) for 5 min, and the supernatant discarded to remove oligonucleotides and other ingredients in a binding buffer that did not bind to E. coli. To remove the oligonucleotides weakly bound to E. coli cells, the cells were resuspended and vortexed at room temperature for 5 min. The cell suspension was recovered by centrifugation (13,000 rpm) at room temperature for 10 min, and the supernatant discarded.
The amount of bound DNA was calculated by UV measurement of the concentration of DNAs washed out in the supernatant after centrifugation. This centrifugation-based partitioning process, including centrifugation, removal of the supernatant, resuspension, and vortexing, was sequentially repeated 10 times. Negative selection was then similarly performed 10 times using a mixture of B. subtilis, K. pneumonia, C. freundii, E. aerogenes, and S. epidermidis (107 cells), as detailed elsewhere. Bound aptamers to E. coli and DNA from E. coli were isolated, and the aptamers were PCR amplified to double-stranded DNA for conventional cloning and sequencing. The secondary structures of several selected aptamers were predicted by free-energy minimization using Zucker’s web-based software Mfold.
The affinity of three aptamers with five stem-loop regions was investigated by binding to target E. coli cells using 3’-FAM dye-labeled aptamers with constant shaking (1000 rpm) at room temperature for 1 h. For the selectivity test, the binding of these labeled aptamers to B. subtilis (shown here), K. pneumonia, C. freundii, E. aerogenes, or S. epidermidis was assessed in a similar manner. Details are available elsewhere.
Results and Discussion

The selectivity of the three 3-FAM labeled aptamers for target (E. coli) vs. non-target (B. subtilis, K. pneumonia, C. freundii, E. aerogenes, and S. epidermidis) bacterial cells used for negative selection showed that the extent of binding to non-target bacteria was less than 25% of that to target E. coli. Based on the affinity and selectivity tests, the aptamers isolated via centrifugation-based partitioning had similar properties to those isolated by the Cell-SELEX method and may therefore be interchangeable for various purposes.
Conclusions
The aforementioned findings obtained with E. coli showed that the centrifugation-based partitioning method allows aptamers to be obtained in a relatively simple and efficient manner compared to conventional Cell-SELEX. In addition, this new method does not require expensive instruments, extensive and time-consuming labor, or a large amount of target cells, which can be a critical issue in some cases. Kim and coworkers state that this method may be general for the other targets, such as proteins or chemicals, if they are appropriately immobilized to a solid matrix that can be easily separable using centrifugation.
Closing Comments
One of the key assumptions of aptamer selection is that the composition of the starting random library is in fact random. Any bias from randomness can cause misrepresentation of “sequence space,” which can in turn limit the search for aptamer sequences that bind the desired target with high specificity. Importantly, the randomness of the TriLink library manufacturing process has been experimentally validated by high-throughput sequencing and bioinformatic analysis through collaborative work by TriLink and NeoVentures, a Canadian aptamer company, as detailed elsewhere.
In the introductory backstory, it was mentioned that aptamers with greater diversity of molecular structure can be prepared by introducing chemical modifications. This can be achieved after SELEX by choosing DNA or RNA aptamers for chemical synthesis, in which various types of chemically modified phosphoramidite reagents are employed to introduce modifications at specific locations. The effect on binding is then measured. Nuclease resistance can also be introduced into aptamers in this manner. Alternatively, structural diversity via chemical modifications can be introduced during SELEX by taking advantage of TriLink's wide selection of modified nucleoside triphosphates, as discussed in a White Paper by TriLink at this link. Recently, the success of this approach has been demonstrated by uber-famous Floyd Romesberg and collaborators, who evolved a polymerase for recognition of 2’-azido-2’-deoxyribonucleoside triphosphates obtained from TriLink (see Methods section in Supporting Information at this link). This enabled the selection of aptamers with large hydrophobic 2′ substituents, which bind human neutrophil elastase or the blood coagulation protein factor IXa.
Finally, it is worth mentioning that on April 21, 2020, NeoVentures announced that it is developing an aptamer for SARS-CoV-2, the recently emerged coronavirus strain responsible for COVID-19.
Your comments are welcomed, as usual.