- MiRNA Biomarker Discovery and Applications Predominate
- Cellular Communication Via MiRNA in Extracellular Vesicles Is Also of Major Interest
- Recent Examples of MiRNA in Cancers and Other Medical Conditions Are Presented
Introduction
Although microRNA (miRNA) and short-interfering RNA (siRNA) were initially discovered in distinct studies, these types of small RNA are closely related in their biogenesis, assembly into RNA–protein complexes, and ability to negatively regulate gene transcripts in diverse eukaryotes, as reviewed elsewhere. Both miRNAs (~21-23 nucleotides) and siRNAs (~21-25 nucleotides) are generated by Dicer (DCR), a multidomain enzyme of the RNase III family. DCR cuts long, double-stranded RNA (dsRNA) into siRNAs and chops short precursor miRNAs (pre-miRNA) with imperfect stem-loop structure into miRNAs.
Exogenous synthetic siRNA is largely of interest because of its ability to down-regulate gene expression through RNA interference (RNAi) for either basic research or nucleic acid-based therapeutics, as featured in several earlier Zone blogs. Human miRNAs are currently estimated to number ~1,100, and they have a major regulatory impact on gene expression through the facilitation of sequence-specific RNAi. Mediated by Argonaute proteins and other components of RISC (RNA-induced Silencing Complex), miRNAs bind complementary sequences within mRNA transcripts, resulting in decreased expression levels of target proteins, as depicted here.
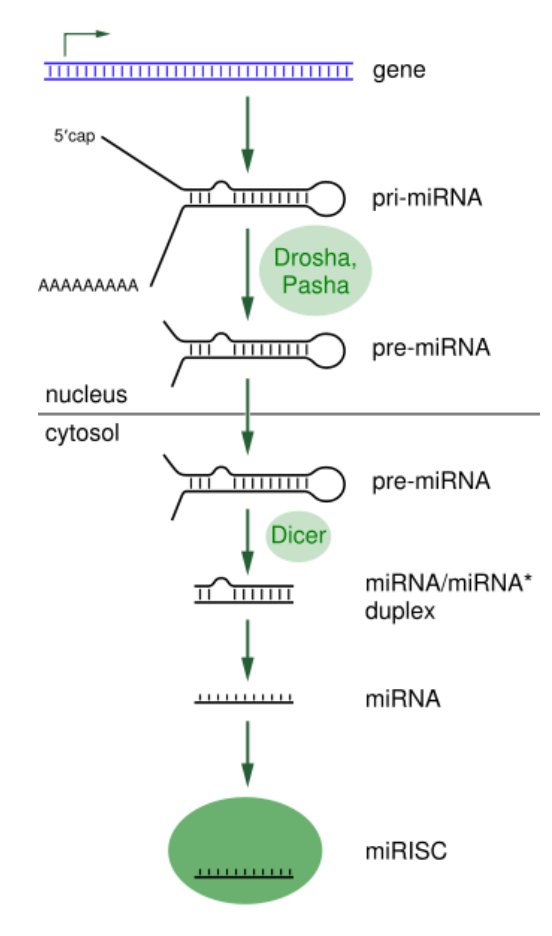 MiRNA processing. Taken from commons.wikimedia.org and free to use.
MiRNA processing. Taken from commons.wikimedia.org and free to use.
Importantly, variations in miRNA levels have been reported in patients’ solid tissues, blood, and other body fluids, making miRNAs promising candidates as biomarkers for many diseases and cellular conditions. Increasing interest in miRNA biomarkers is evidenced by the chart of publications indexed in the NIH PubMed database. Of the various methods available for the quantification of miRNA, sequencing is the only one with the added advantage of being able to be utilized for the discovery of candidate miRNAs without prior knowledge of the miRNA sequences.
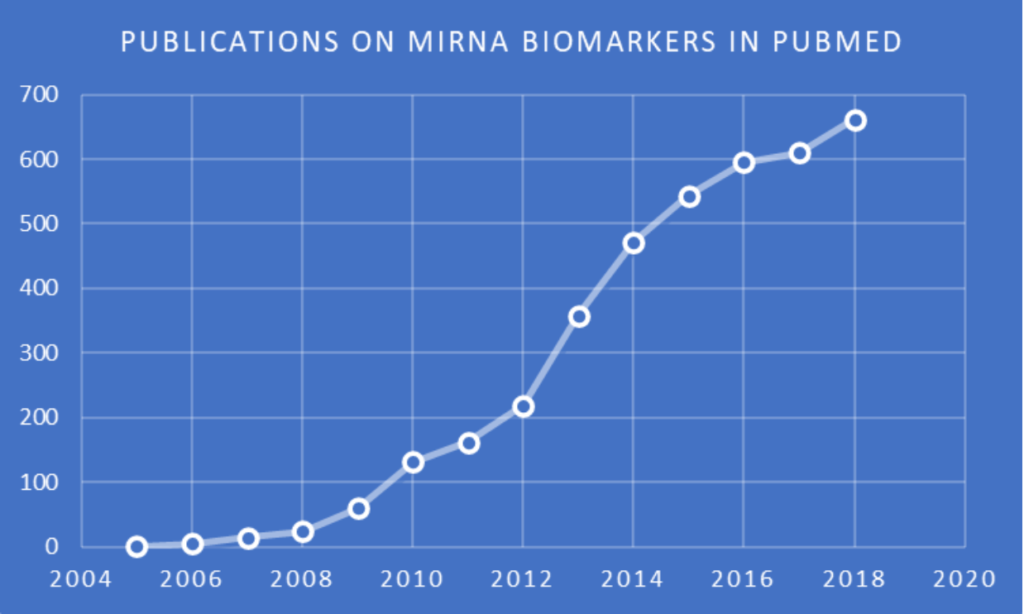 Publications in PubMed indexed to (biomarker OR biomarkers) AND miRNA in the title and/or abstract. Search and chart by Jerry Zon.
Publications in PubMed indexed to (biomarker OR biomarkers) AND miRNA in the title and/or abstract. Search and chart by Jerry Zon.
The sequencing of miRNA is carried out by first preparing a pool (aka library) of all small RNAs (sRNAs) in a sample for conversion to complementary DNA (cDNA) for high-throughput (aka next-generation) sequencing (NGS). This is usually achieved by ligation of the sRNA pool to adapters for reverse transcription into cDNA and amplification. With the development of the CleanTag® Small RNA Library Preparation Kit, researchers at TriLink were also able to minimize sequence-based bias and adapter dimer formation, as depicted here.
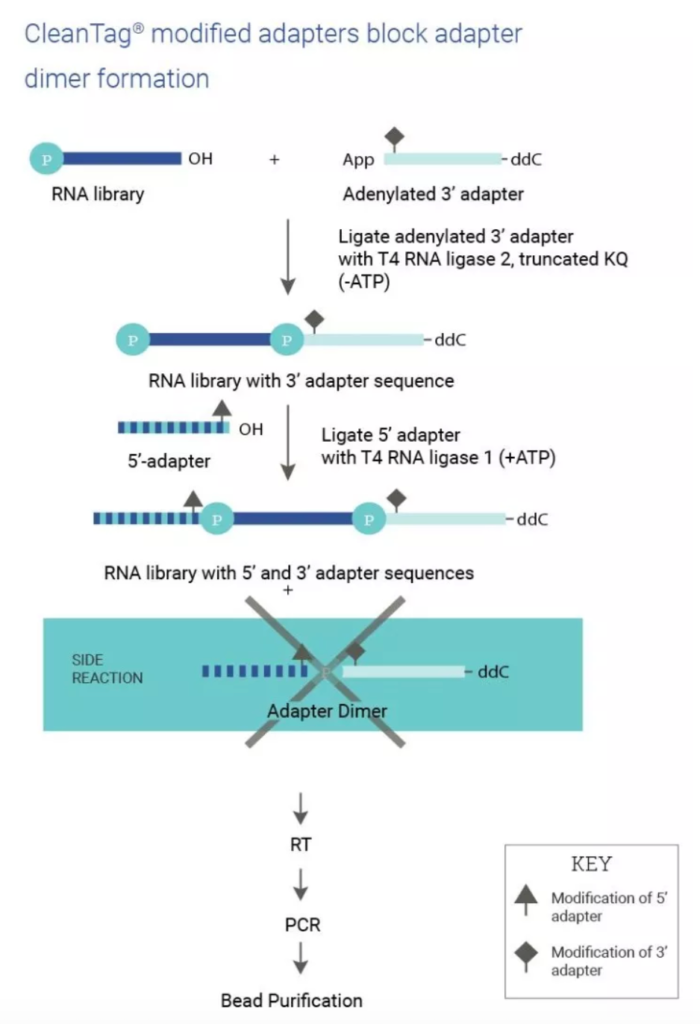 Taken from TriLink Biotechnologies.
Taken from TriLink Biotechnologies.
For details on the initial development and improved performance of CleanTag® technology, interested readers can follow this link to a research article by Shore et al. in PLOS ONE that has had more than 13,000 views since its online publication in November 2016. After this work, a more detailed chapter in Next-Generation Sequencing Methods and Protocols was released, followed by a collaborative publication on two different methods for single-cell (sc)-lysis coupled with CleanTag®. These methods enable sc-sRNA-seq and offer a relatively short hands-on time of 6 hours, while also showing detection of miRNA with various abundances, ranging from 16,000 copies/cell to about 10 copies/cell.
The next sections will provide examples of selected 2018-2020 publications by investigators who used CleanTag® for sRNA-seq in the study of miRNA as biomarkers or for other medically related purposes.
Extracellular Vesicles

Cells release various types of membrane vesicles into the extracellular environment. These can range in size from 100-1000 nm, and are called exosomes when derived from endosomal membranes, or microvesicles when derived from plasma membranes. As reviewed elsewhere, these extracellular vesicles (EVs) represent an important mode of intercellular “communication,” serving as vehicles for transfer between cells of membrane and cytosolic proteins, lipids, and RNA, especially translatable mRNA and post-translation regulatory miRNA. Unfortunately, the lack of knowledge on the molecular mechanisms for EV formation, as well as the fact that there are no methods available to interfere with the packaging of cargo or vesicle release, means that the physiological relevance of extracellular vesicles in vivo remains a mystery. In this review, we focus on the characterization of EVs and on the currently proposed mechanisms of their formation, targeting, and function.
By searching the term CleanTag® in Google Scholar, 13 publications on EVs were found to use CleanTag® for sequencing. Of these reports, the Zone selected four to briefly discuss the results obtained.
Amyotrophic Lateral Sclerosis (ALS): Patients with this neurodegenerative disorder have a mean survival of 2 to 5 years. Early and efficient diagnosis of the various forms of ALS remains a significant challenge, which makes the identification of clinically-relevant biomarkers in readily accessible body fluids even more essential. Saucier et al. investigated whether an ALS-associated miRNA signature in EVs, which can cross the blood-brain barrier and enter the circulatory system, can be found in plasma samples of people diagnosed and living with ALS (PALS). Compared to healthy control subjects, NGS revealed elevated levels of 5 miRNAs and reduced levels of 22 miRNAs in EVs collected from PALS. MiRNAs with relevance to ALS were found to be deregulated. It was concluded that “[t>
hese data identify an ALS-associated miRNA signature in EVs of PALS and further strengthen the potential diagnostic relevance of these small molecules for this condition.”


Neonatal Pulmonary Hypoplasia (PH): According to Antounians et al., incomplete lung development, also known as pulmonary hypoplasia (PH), is a recognized cause of neonatal death. There is no effective treatment that promotes fetal lung growth and maturation, and to begin to fulfill this unmet need, researchers developed a stem cell-based approach that enhances fetal lung development via the administration of EVs derived from amniotic fluid stem cells (AFSCs) in rat models of PH. The miRNAs present in AFSC-EVs were shown to be responsible for these beneficial effects. In brief, administration of AFSC-EVs increased terminal bud density and surface area of lung explants back to control levels, as well as promoted lung epithelial cell differentiation in lung organoids. It was stated that the AFSC-EVs contain 901 miRNAs, some of which are crucial for fetal lung development, such as miR17 ~ 92 cluster, and that “AFSC-EVs represent a promising therapeutic strategy for PH in fetuses.”
Infant Congenital Heart Disease (CHD): According to Trac et al., CHD affects an estimated 40,000 infants each year in the United States. Stem cell therapy, including c‐kit+ progenitor cells (CPCs), is currently being studied as a potential treatment for right ventricular heart failure (RVHF) occurring in complex forms of CHD. Researchers studied the contents of pediatric progenitor cell exosomes in conjunction with existing computational modeling. Briefly, they demonstrated that the top 40 miRNAs from CPC exosomes can be related to in vitro angiogenesis and antifibrosis responses by mathematical modeling. Although the model was trained on in vitro data, it predicted angiogenesis and fibrosis responses to exosome treatment in vivo with a strong correlation to published in vivo responses. It was concluded that this modeling of exosome miRNA content has “the potential to inform preclinical trials and predict new promising CPC therapies.”
Cancers

Liver Cancer: Fibrolamellar carcinoma (FLC) is a rare liver cancer that primarily affects adolescents and young adults. According to Dihn et al., the role of miRNAs in FLC remains unclear, which led to the investigation of whether dysregulated miRNAs in FLC contributes to FLC pathogenesis. Briefly, sRNA-seq was performed on a FLC patient-derived xenograft model, and on purified cell populations of the liver to determine whether key miRNA changes were tumor cell-intrinsic. The researchers identified miR-375 as the most dysregulated miRNA in primary FLC tumors (27-fold down-regulation), and its expression was also significantly decreased in the FLC patient-derived xenograft model compared to 4 different cell populations of the liver. Finally, they showed that overexpression of miR-375 mitigated tumor cell growth and invasive potential. It was concluded that these findings open a potentially new molecular therapeutic approach for the treatment of FLC.
Brain Tumors: According to Kopkova et al., the annual incidence rate of primary brain tumors and brain metastases is ~40 for every 100,000 people in the world, and growing. As prognosis and therapy differ between brain tumor types, accurate and early diagnosis may improve survival and quality of life for these patients. However, current diagnostic approaches are limited by the localization and tissue heterogeneity of brain tumors. Cerebrospinal fluid (CSF) bathes the central nervous system (CNS) and is considered to reflect all pathological conditions. From this perspective, CSF could be an ideal source of diagnostic biomarkers for brain tumors.

The researchers carried out sRNA-seq of 89 CSF samples taken from 32 glioblastoma (GBM), 14 low-grade glioma (LGG), 11 meningioma, and 13 brain metastasis patients, as well as from 19 non-tumor donors. NGS analysis revealed 25, 3, 2, and 14 CSF miRNAs were significantly differentially expressed in GBM, meningiomas, LGG, and metastasis patients, respectively, compared to control CSF samples. In addition, 6 miRNAs showed different levels between GBM and LGG. It was concluded that there is potential of utilizing CSF miRNAs as useful biomarkers for brain tumors. Subsequently published sRNA-seq analyses of additional CSF samples have confirmed and extended these conclusions.
Rectal Cancer: Machackova et al. state that rectal cancer accounts for approximately one-third of all colorectal cancers. Currently, the standard treatment for locally advanced rectal cancer (LARC) is neoadjuvant chemoradiotherapy (CRT) with capecitabine or 5-fluorouracil, followed by curative surgery. Unfortunately, only 20% of patients with LARC present complete pathological response after CRT, and the response is poor or absent in 20-40% cases. The researchers investigated whether miRNAs in tumor biopsy specimens have the potential to predict therapeutic response in LARC patients.
A total of 87 LARC patients treated by CRT were enrolled in a prospective study, in which sRNA-seq was used for 40 tumor biopsy samples of LARC patients (20 responders, 20 non-responders) and qPCR validation of selected miRNA candidates. In the discovery phase of the study, 69 miRNAs were identified as having significantly different expression among the two groups. Among these miRNAs, 48 showed a lower expression and 21 showed higher expression in tumor tissues from poorly responding LARC patients. Five miRNAs were selected for validation, but only miR-487a-3p was confirmed to have a significantly higher expression in the tumor biopsy specimens of non-responders to neoadjuvant CRT (p<0.0006, AUC=0.766). Gene Ontology (GO) clustering and pathway enrichment analysis of the miR-487a-3p mRNA targets revealed potential mechanisms behind miR-487a-3p roles in resistance to CRT. It was concluded that miR-487a-3p is a potential predictive biomarker of CRT response in LARC patients.

Leukemia: According to Assmann et al., TCRαβ+ CD8+ large granular lymphocyte (LGL) leukemia is a rare, chronic, heterogeneous and hematological disorder that mostly affects the elderly. Although the precise disease etiology is largely unknown and disease course is heterogeneous, it is generally accepted that LGL cells arise through chronic antigenic stimulation and inflammation, thriving due to the dysregulation of proliferation and apoptosis pathways. One level of dysregulation of these pathways could involve miRNAs. The investigators explored the “miRNAome” of this particular type of LGL leukemic cells by performing sRNA-seq on sorted TCRαβ+ CD8+ LGL cells (n = 7) and sorted age-matched healthy control cells (n = 6). Furthermore, they evaluated whether differentially expressed miRNAs could potentially drive the dysregulation of proliferation and apoptosis pathways in LGL leukemia.
Principle component analysis of miRNA expression profiles clearly showed LGL leukemia cells clustering away from healthy control T cells, with more than 100 miRNAs significantly upregulated or downregulated between patient and control cells. Pathway analysis of top differentially expressed miRNAs indicated that these miRNAs could be involved in cell-cycle regulation through repression of cyclin dependent kinases (CDKs) in the NF-κB pathway, while also potentially driving apoptosis resistance through inhibition of the p53 and BCL2 pathways.
Crohn’s Disease

MiRNAs were screened in feces of 52 patients with CD and 15 healthy controls using sRNA-seq. The results were confirmed by PCR. The relationship between fecal miRNA levels and the clinical CD activity index (CDAI) or CD endoscopic index of severity (CDEIS) was explored, respectively. MiR-192-5p, miR-375, and miR-141-3p correlated with both CDAI and CDEIS, whereas miR-15a-5p correlated only with CDEIS. It was concluded that these fecal miRNAs show promise as biomarkers for monitoring CD progression.
In another study of CD published by Visvanathan et al., Global transcriptome-wide smiRNA-seq of biopsy samples from 40 patients with colonic or ileocolonic CD receiving treatment with risankizumab, a monoclonal antibody targeting the IL-23 p19 subunit, identified 18 significantly differentially expressed miRNAs versus placebo at week 12. Of these, miR-223-3p was among the greatest downregulated miRNAs in the colon and in feces.
Osteoporosis: Kocijan et al. state that miRNAs control the activity of a variety of genes crucial to bone metabolism and, therefore, the clinical utility of miRNAs as biomarkers and drug targets for bone diseases merits further investigation. To address this possibility, the researchers investigated the use of an animal model of postmenopausal osteoporosis to generate a comprehensive dataset on miRNA regulation in bone tissue and peripheral blood during bone loss, specifically with regards to anti-resorptive and osteo-anabolic treatment.

Forty-two rats were randomized to “sham” surgery (n = 10) or ovariectomy (OVX, n = 32). Eight weeks after surgery, OVX animals were further randomized to anti-resorptive treatment with zoledronate (n = 11), osteo-anabolic treatment with teriparatide (n = 11), a parathyroid hormone, or vehicle treatment (n = 10). After 12 weeks of treatment, bone and serum samples were used for miRNA analysis by sRNA-seq, mRNA levels using RT-qPCR, and bone microarchitecture analysis using nano-computed tomography (nano-CT) scans.
Ovariectomy resulted in loss of trabecular bone, which was fully rescued using osteo-anabolic treatment, and partially rescued using anti-resorptive treatment. NGS revealed that both anti-resorptive and anabolic treatment had a significant impact on miRNA levels in bone tissue and serum.
Out of 426 detected miRNAs, 46 miRNAs were regulated by teriparatide treatment and 10 by zoledronate treatment. Interestingly, teriparatide and zoledronate were able to revert miRNA changes in tissue and serum of untreated OVX animals, such as the up-regulation of miR-203a-3p, a known osteo-inhibitory miRNA. Researchers confirmed previously established mechanisms of miR-203a by analyzing its direct target Dlx5 in the femoral head. It was concluded that the observed effects of bone loss and treatment response on miRNA levels in bone are also reflected in the peripheral blood, suggesting the possibility of minimally-invasive monitoring of bone-derived miRNAs using liquid biopsies.
Concluding Comments

Your comments are welcomed, as usual.







