- Envisaged in the 2002 Challenge for Achieving a $1,000 Genome
- Are We There Yet? Yes…and No
- So Where Are We, Actually?
The notion of metaphorically ‘dirt cheap’ genome sequencing is now so prevalent that it seems to have been always available—and have virtually unlimited utility—as previously touted in a provocative article in Nature Biotechnology rhetorically entitled What would you do if you could sequence everything? The notion of ‘everything’ obviously includes all people, and—as we’re reminded by the now familiar t-shirt statement—‘babies are people too.’ Unlike the ‘big bang’ origin of the universe, cheap-enough-sequencing-for-everything, including all babies (newborns, actually) just didn’t happen spontaneously, so when and how did this come about?
In the Beginning…
According to the bible, the history of creation began when, “in the beginning God created the heavens and the earth.” The history of cheap-enough-sequencing-for-everything, according to my tongue-in-cheek reckoning, is that “In the beginning Craig Venter convened and moderated a diverse panel of experts to discuss The Future of Sequencing: Advancing Towards the $1,000 Genome.” This was a ‘hot topic’ at the 14th International Genome Sequencing and Analysis Conference (GSAC 14) in Boston on Oct 2nd 2002. The pre-conference press release added that “the panel will explore new DNA sequencing technologies that have the potential to change the face of genomics over the next few years.”
While it’s taken considerably more than a ‘few years’ to achieve the $1,000 genome, continual decrease in sequencing costs has enabled large-scale sequencing initiatives such as the 1000 Genomes Project. Nick Loman’s 2013 blog entitled The biggest genome sequencing projects: the uber-list! outlines the 15 largest sequencing projects to date and takes a look at some of the massive projects that are currently in the works.
According to GenomeWeb on Jan 14th 2014, Illumina launched a new sequencing system that can now produce a human genome for under $1,000, claiming to be the first to achieve this ‘long sought-after goal’—roughly 12 years in the making, by my reckoning. A LinkedIn post on Jan 15th by Brian Maurer, Inside Sales at Illumina, quotes Illumina’s CEO as saying that "one reagent kit to enable 16 genomes per run will cost $12,700, or $800 per genome for reagents. Hardware will add an additional $137 per genome, while sample prep will range between $55 and $65 per genome." While Illumina is staking claim to the $1,000 genome and customers acknowledge the new system provides a drastic price reduction, the actual costs of sequencing a genome are being debated. Sequencing service provider, AllSeq, discusses the cost breakdown in their blog. While initially a bit negative, their outlook on the attainable costs seems to be improving.
So, with affordable genome sequencing apparently being a reality, are we ready to sequence every newborn?
Yes…and No
Why this conflicting answer of ‘yes…and no’? The ‘yes’ part is based on the fact that NIH has recently funded four studies on the benefits—and risks—of newborn genome sequencing. The ‘no’ part reflects the added facts that these studies will take five years, and will look at how genome testing of newborns could improve screening, and address what some geneticists view as their most sensitive ethical questions yet.
Put another way, and as detailed in the following section, the requisite low-cost DNA sequencing technology is now available but it needs to be demonstrated—through technical feasibility investigations and in clinical pilot studies—that newborns receive health benefits of the type expected, and that numerous ‘tricky’ ethical issues can be dealt with in an ‘acceptable manner’—although to whom it is acceptable is not obvious to me at this time. Likely controversial views on reimbursement also have to be addressed, but that’s a whole other topic—dare I say ‘political football’—vis-à-vis Obamacare, ooops, I mean the Affordable Care Act.
So What’s the Plan?
The following answer is an adaptation of the Sep 13th 2013 Science News & Analysis by Jocelyn Kaiser entitled Researchers to Explore Promise, Risks of Sequencing Newborns’ DNA.
The National Institute of Child Health and Human Development (NICHD) is rolling out a $25 million, 5-year federal research program to explore the potential value of looking at an infant’s genome to examine all of the genes or perhaps a particularly informative subset of them. This genome testing could significantly supplement the decades-old state screening programs that take a drop of blood from nearly every newborn's heel and test it for biochemical markers for several dozen rare disorders. Diagnosing a child at birth can help prevent irreversible damage, as in phenylketonuria, a mutant-gene metabolic disorder that can be controlled with diet.
 Handle with care. Genome testing could enhance newborn screening, but it raises ethical issues [credit: Spencer Grant/Science Vol. 341, p. 1163 (2013)>
Handle with care. Genome testing could enhance newborn screening, but it raises ethical issues [credit: Spencer Grant/Science Vol. 341, p. 1163 (2013)>
.
The ethical concern here is that genome sequencing, unlike the current newborn screening tests, could potentially reveal many more unexpected genetic risks, some for untreatable diseases. Which of these results should be divulged is already controversial, according to Kaiser, who added that “sparks are still flying” over an earlier report described in Science as follows:
Geneticists, ethicists, and physicians reacted with shock to recommendations released last week by the American College of Medical Genetics and Genomics: that patients undergoing genomic sequencing should be informed whether 57 of their genes put them at risk of serious disease in the future, even if they don't want that information now. The recommendations also apply to children, whose parents would be told even if illness wouldn't strike until adulthood. The advice runs counter to the long-standing belief that patients and parents have the right to refuse DNA results. This is the first time that a professional society has advised labs and doctors what to do when unanticipated genetic results turn up in the course of sequencing a patient's genome for an unrelated medical condition.
Given this background, it’s reassuring—in my opinion—that NICHD is taking a ‘go slow’ approach. I’m further reassured that NICHD is funding research of technical and ethical/social issues in four different but interrelated studies (see table below). Kaiser adds that “all will examine whether genomic information can improve the accuracy of newborn screening tests, but they differ in which additional genes they will test and what results they will offer parents.”
 New ground: four projects funded at a total of $25 million over 5 years will look at how genome testing could improve newborn screening and other questions [credit: Spencer Grant/Science Vol. 341, p. 1163 (2013)>
New ground: four projects funded at a total of $25 million over 5 years will look at how genome testing could improve newborn screening and other questions [credit: Spencer Grant/Science Vol. 341, p. 1163 (2013)>
.
A team at the University of North Carolina is studying how to return results to low-income families and others who might not be familiar with genomics. It is also dividing genetic findings into three categories—mutations that should always be reported; those that parents can choose to receive, which might include risk genes for adult cancers; and a third set that should not be disclosed, e.g. untreatable adult-onset diseases such as Alzheimer's.
A team at Brigham and Women's Hospital in Boston and Boston Children's Hospital hopes to learn how doctors and parents will use genomic information. ‘We're trying to imagine a world where you have this information available, whether you're a sick child or healthy child. How will it change the way doctors care for children?’ asks co-principal investigator Robert Green.
Ethicist Jeffrey Botkin of the University of Utah opined that sequencing might never replace existing newborn screening because of its costs and the complexity, according to Kaiser. However, Kaiser said that Botkin and others believe that it’s important to explore these issues because wealthy, well-informed parents will soon be able to mail a sample of their baby's DNA to a company to have it sequenced—regardless of whether medical experts think that's a good idea. ‘There's an appetite for this. It will be filled either within the medical establishment or outside of it,’ Kaiser quotes Green as saying.
I should add that this parental ‘appetite’ doesn’t seem to be easily satisfied at the moment, based on my—admittedly superficial—survey of what’s currently available in the commercial genome sequencing space. For now, companies such as Personalis and Knome restrict their offerings to researchers and clinicians, not direct-to-consumers, such as parents-on-behalf-of-newborns—yet.
Sample-to-Whole-Genome-Sequencing-Diagnosis in Only 50 Hours
Having been a laboratory investigator during the stunning evolution from manual Maxam-Gilbert sequencing to highly automated Sanger sequencing—the title of this section ‘blows my mind’ and seems impossible—but it’s not! Stephen Kingsmore and collaborators at the Children's Mercy Hospital in Kansas City, Missouri reported this remarkable achievement in Science Translational Medicine in 2012 and, as noted in the aforementioned table, aim to cut this turnaround time to within 24 hours!
In that 2012 report, they make a compelling case for whole-genome sequence-based diagnostics—and super speedy sample-to-result by noting that monogenic diseases are frequent causes of neonatal morbidity and mortality, and disease presentations are often undifferentiated at birth. Of the ~4,000 monogenic diseases that have been characterized, clinical testing is available for only a handful of them and many feature clinical and genetic heterogeneity. Hence, an immense unmet need exists for improved molecular diagnosis in infants. Because disease progression is extremely rapid—albeit heterogeneous—in newborns, molecular diagnoses must occur quickly to be relevant for clinical decision-making.
Using the workflow and timeline outlined below, they describe 50-hour differential diagnosis of genetic disorders by whole-genome sequencing (WGS) that features automated bioinformatics analysis and is intended to be a prototype for use in neonatal intensive care units. I should add that an automated bioinformatics analysis is critical for clinical utility, and has been the subject of ‘musings’ by Elaine Mardis in Genome Medicine entitled The $1,000 genome, the $100,000 analysis?
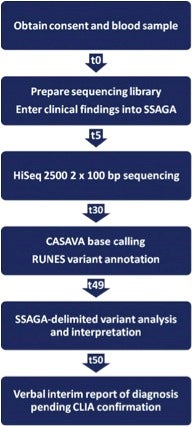 Summary of the steps and timing (t, hours) resulting in an interval of 50 hours between consent and delivery of a preliminary, verbal diagnosis [taken from Saunders et al. Sci Transl Med 4, 154ra135 (2012).
Summary of the steps and timing (t, hours) resulting in an interval of 50 hours between consent and delivery of a preliminary, verbal diagnosis [taken from Saunders et al. Sci Transl Med 4, 154ra135 (2012).
To validate the feasibility of automated matching of clinical terms to diseases and genes, they entered retrospectively the presenting features of 533 children, who have received a molecular diagnosis at Children’s Mercy Hospital within the last 10 years, into symptom- and sign-assisted genome analysis (SSAGA)—a new clinico-pathological correlation tool that maps the clinical features of 591 well-established, recessive genetic diseases with pediatric presentations to corresponding phenotypes and genes known to cause the symptoms. Sensitivity was 99.3%, as determined by correct disease and affected gene nominations.
Rapid WGS was made possible by two innovations. First, a widely used WGS platform was modified to generate up to 140 Gb [Gb = giga base pairs = 1,000,000,000 base pairs>
of sequence in less than 30 hours (Illumina HiSeq 2500). Secondly, sample preparation took 4.5 hours, while 2 × 100 base pairs of genome sequencing took 25.5 hours. The total ‘hands-on’ time for technical staff was 5 hours.
Readers who are interested in more technical details for sample prep, sequencing, and bioinformatics/analytics should read the full text. However, the authors’ abstract provides the following succinctly described diagnostic ‘payoff’, so to speak:
Prospective WGS disclosed potential molecular diagnosis of a severe GJB2-related skin disease in one neonate; BRAT1-related lethal neonatal rigidity and multifocal seizure syndrome in another infant; identified BCL9L as a novel, recessive visceral heterotaxy gene (HTX6) in a pedigree; and ruled out known candidate genes in one infant. Sequencing of parents or affected siblings expedited the identification of disease genes in prospective cases. Thus, rapid WGS can potentially broaden and foreshorten differential diagnosis, resulting in fewer empirical treatments and faster progression to genetic and prognostic counseling.
These are compelling results, in my opinion. Let me know if you also find this compelling. As always, your comments are welcomed.
Postscript
After finishing the above post, Andrew Pollack at The New York Times published a fascinating article giving examples of how pharmaceutical companies are heavily investing in genetic studies that employ exome-sequencing of large study groups in a search for clues to aid drug development. Regeneron is conducting one such study that includes 100,000 genomes.






