- Modified mRNA (Mod-RNA) for Therapeutics Drives Development of New Technologies
- Among These Is Tagging Mod-RNA for Visualization In Vivo
- Tagging Mod-RNA with TAG Earns Neal Devaraj the 2018 Blavatnik National Award in Chemistry
Back in 2013, I wrote a blog titled Modified mRNA Mania, which drew attention to the then nascent development of an entirely new field of therapeutics, based on the use of biosynthetic modified mRNA (mod-RNA). I also wrote a more recent blog about this still trending field, titled Deluge of mRNA Delivery Publications, featuring a chart of supporting publication data to emphasize the exponentially growing efforts to improve delivery.
 Prof. Neal Devaraj. Credit Neal Devaral. With permission
Prof. Neal Devaraj. Credit Neal Devaral. With permission
The present blog in this ongoing series on mod-RNA focuses on in vivo visualization of mod-RNA by use of a very clever enzyme-mediated labeling method. Importantly, this labeling procedure can be applied to any biosynthetic mod-RNA of interest. The method was invented by Neal Devaraj, an Associate Professor of Chemistry and Biochemistry at the University of California, San Diego. Because of his work, Prof. Devaraj recently received the 2018 Blavatnik National Award in Chemistry, which you can read about at the end of this blog.
Backstory for Tagging Mod-RNA
In vivo expression of therapeutic proteins encoded in exogenous mRNA provides several advantages compared to delivery of the corresponding complementary DNA. Delivering mRNA is easier because it only needs to enter the cytoplasm, rather than the nucleus, to be functional (depicted here in scheme B). This avoids complications from transcription regulation machinery. Additionally, mRNA does not permanently alter the genome like genomically integrated DNA does (depicted here in scheme A), therefore avoiding permanent and potentially lethal changes.
 Taken from Avci-Adali et al. J. Biol. Eng. 2014, 8, 8. Copyright © BioMed Central Ltd. With permission.
Taken from Avci-Adali et al. J. Biol. Eng. 2014, 8, 8. Copyright © BioMed Central Ltd. With permission.
In order to overcome the intrinsically unstable and transient nature of natural RNA, the incorporation of modified nucleotides has been—and continues to be—explored to harness RNA as a therapeutic agent. Multiple studies have focused on providing an increase in serum stability, a less active immune response, and an increase in translational capacity, as reviewed elsewhere.
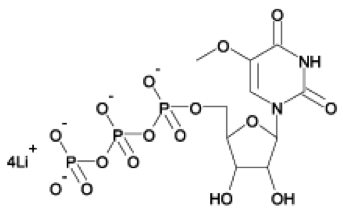 5-moUTP lithium salt. Taken from TriLink BioTechnologies
5-moUTP lithium salt. Taken from TriLink BioTechnologies
Many types of modified nucleobases can be incorporated into mRNA transcripts by substituting natural nucleotide triphosphates with one or more modified nucleotide triphosphates during enzymatic synthesis with an RNA polymerase. TriLink BioTechnologies R&D group has synthesized and screened numerous modified nucleotide triphosphates, and 5-methoxyuridine (5moU) has been shown to be especially useful. Interested readers can learn more about mod-RNA at this link to a video presentation in 2018 by Dr. Anton McCaffrey, Senior Director of Emerging Science and Innovation at TriLink.
In order to fully exploit the therapeutic potential of mod-RNAs, novel strategies for the safe and effective “decoration” or tagging of in vitro transcribed RNA, as well as functional moieties or fluorophores to allow for visualization, must be developed. A convenient bioconjugation method that is capable of appending targeting molecules or fluorophores to therapeutic Mod-RNAs would enable new modalities of highly specific uptake of RNA through endocytic pathways, as well as visualization.
Tagging Mod-RNA with TAG
To address the need for a specific and convenient bioconjugation method for mod-RNA, Devaraj and his students envisioned decorating mod-RNA by use of an RNA modifying enzyme. In their 2018 publication in Molecular Pharmaceutics, it was noted that more than 100 RNA post-transcriptional modifications have been reported to date, with a large majority of these modifications occurring on transfer RNAs (tRNAs). Among these, bacterial tRNA guanine transglycosylases (TGTs) have been extensively studied. As depicted here, during the transglycosylation reaction, the N−C glycosidic linkage of a key guanine at the wobble position of the anticodon loop is cleaved, and a 7-deazaguanine derivative named preQ1 (R = H) is substituted.

Importantly, the RNA-bound TGT crystal structure previously reported by others suggested to Devaraj that there might be enough room to chemically modify the natural preQ1 (R = H) substrate and thus repurpose the enzyme to covalently append a larger R group, such as a fluorophore or affinity tag. This concept was initially reduced to practice in a preliminary communication, wherein the minimal 17-nucleotide hairpin sequence shown above, which was named TAG, was genetically engineered into the 3′-UTR of a full mRNA transcript coding for the red fluorescent protein mCherry (mCherry-TAG). When treated with bacterial TGT and a preQ1 derivative with R = dye, fluorescence-based gel analysis confirmed the formation of the expected mCherry-TAG-dye conjugate.
In a recent Molecular Pharmaceutics publication, Devaraj and his students extended this E. coli TGT activity to recognize TAG in mCherry mod-RNA comprised of the chemically modified nucleobases 5-methylcytosine (5mC) or pseudouracil (Ψ), as well as doubly substituted 5mC + Ψ mod-RNA. The approach depicted here is a two-step process involving RNA-TAG covalent conjugation followed by tetrazine biorthogonal labeling.

In vitro transcription (IVT) reactions for mCherry-TAG mod-mRNA transcripts were performed with both a partial (25%) and a complete (100%) replacement of the natural bases with 5mC and Ψ by use of the corresponding modified triphosphates, which I’m pleased to say were obtained from TriLink. A cap analog was added to the 5’ end and, after IVT, the 3’ end was polyadenylated to furnish mature mRNAs capable of being translated into mCherry-TAG upon transfection into mammalian cells.
Although mRNA-TAG has been established to efficiently and directly incorporate small molecule moieties, Devaraj and coworkers state that they envisioned a two-step labeling approach of mod-mRNA such that the degree of labeling could be more concretely proven. Additionally, this methodology would also allow a diverse array of targeting molecules, irrespective of size or class, to be easily conjugated and evaluated without the need for multiple syntheses of preQ1 derivatives or the need to quantify each substrate’s efficiency of incorporation.
As depicted here, appending a bioorthogonal tetrazine moiety to mod-mRNA transcripts (red) can be followed by coupling a trans-cyclooctene (TCO)-functionalized fluorescent probe or affinity agent (blue) using previously established, robust tetrazine-ligation chemistry, which is reviewed elsewhere.

The mCherry mod-mRNAs were treated with the TGT enzyme at 37 °C for 4 h to first access tetrazine-labeled mCherry mod-mRNAs. The purified tetrazine-conjugated mod-RNA was subsequently conjugated to a TCO-functionalized sulfo-Cy5 fluorescent probe via tetrazine ligation by incubating for 4 h at 37 °C. The purified RNAs were subjected to polyacrylamide gel electrophoresis and imaged. As shown here, the degree of labeling (DOL) was determined to be ~95% for unmodified transcript, ~84% for 5mC transcript, ~66% for Ψ modified transcript, and ~61% for both Ψ and 5mC modified transcript.

Devaraj and coworkers hypothesize that the slightly lowered efficiency in labeling of the all 5mC-mod-mRNAs in comparison to unmodified mRNA could be due to variations in the secondary structure of the RNA-TAG hairpin, which may hamper enzyme substrate recognition. More interestingly, and with reference to the TAG hairpin shown above, they further speculate that the decrease in DOL for the Ψ containing mod-mRNA transcripts most likely arises from disruption of key interactions between the TGT enzyme and flanking uridine residues on either side of the exchanged guanine when substituted for the modified base Ψ.
Conclusion
Devaraj and coworkers concluded that they have developed a relatively simple methodology to covalently label mod-mRNAs with a modular two-step approach that can incorporate small molecules such as imaging agents, affinity handles, targeting agents, and drug conjugates using RNA-TAG. They also concluded that this approach provides the possibility of diverse decoration of therapeutically relevant mod-RNAs, without the limitation of length characteristic of synthetic RNA strategies. Finally, they expressed belief that “the RNA-TAG technology could greatly expand the arsenal of therapeutic RNAs by way of conjugation to a variety of functional decorations relevant to further tuning the emerging modality of RNA as a therapeutic class.”
I fully agree with these conclusions, and welcome your comments, as usual.
Footnote
This past June, The Blavatnik Family Foundation and the New York Academy of Sciences announced the 2018 Laureates of the Blavatnik National Awards for Young Scientists, who will each receive $250,000: the largest unrestricted scientific prize offered to America’s most promising faculty-level scientific researchers 42 years of age and younger. Nominated by 146 research institutions across 42 states, the 286 nominees were narrowed to a pool of 31 Blavatnik National Finalists. From this pool of Finalists, a distinguished scientific jury chose three outstanding Laureates, one in each of the Awards’ scientific disciplinary categories: Life Sciences, Physical Sciences & Engineering, and Chemistry.
Life Sciences: Janelle Ayres, PhD, of the Salk Institute for Biological Studies, for her pioneering research in immunology and the study of how bacteria interact with humans. Dr. Ayres’s work is revolutionizing our understanding of host-pathogen interactions and has the potential to solve one of the greatest current public health threats: anti-microbial resistance.
Physical Sciences & Engineering: Sergei V. Kalinin, PhD, of Oak Ridge National Laboratory, for creating novel techniques to study, measure, and control the functionality of nanomaterials at the atomic and nanoscale levels. Dr. Kalinin’s work manipulating individual atoms has the potential to enable scientists to create new classes of materials by assembling matter, atom-by-atom.
Chemistry: Neal K. Devaraj, PhD, of the University of California, San Diego, for his transformative work on the synthesis of artificial cells and membranes, thus creating an exciting new field of research that aims to address one of the great challenges in synthetic biology. Dr. Devaraj has made several game-changing discoveries, and has pioneered the development of new methods for labeling biological molecules, which have already been adopted by researchers globally.
Here is a link to all of Neal Devaraj’s publications currently indexed in PubMed.






