- The ‘Guardian of The Genome’ p53 Tops the List of Top 10 Genes
- Cancer-Related Genes Are Prevalent Among the Top 10
- TriLink Is Proud of Its Products Mentioned in Studies of the Top 10
People seem to be fascinated with “Top 10” lists, which currently exceed 134,000 according to The Top Tens® website. Later, you can search this mind-boggling list for whatever strikes your fancy; for now, let’s focus on what’s trending in nucleic acids which is the intent of this blog. In that regard, and linked to the notion of “Top 10” lists, venerable Nature magazine recently published a news piece titled The Greatest Hits of the Human Genome. This caught my attention as definitely a blog-worthy topic, as it had a tag line offering readers “a tour through the most studied genes in biology,” which sounded very intriguing to me.
Interested readers can peruse the entire news item in Nature, but for the moment here’s my synopsis of Nature’s list of Top 10 Genes, along with my findings for representative examples of how TriLink products have been used for each of these Top 10 Genes.
The Top 10 Genes
According to Nature, software engineer pre-doctoral student Peter Kerpedjiev became intrigued to rank the world’s most studied human genes, and eventually collaborated with Nature in analyzing data extracted from the NCBI/NLM PubMed database. Following is the rank-order list of the Top 10 most studied genes, along with a brief functional definition of each gene, and then a graphical depiction of the number of citations for each gene. Incidentally, you’ll see that naming genes is not systematic, and simply adopts letter and number abbreviations used in the early literature as descriptors.
- TP53 Tumor-suppressor protein p53 is mutated in up to half of all cancers.
- TNF Tumor necrosis factor has been a drug target for cancers and inflammatory diseases.
- EGFR Epidermal growth factor receptor is a membrane-bound protein often mutated in drug-resistant cancers.
- VEGFA Vascular endothelial growth factor A promotes growth of blood vessels.
- APOE Apolipoprotein E has important roles in cholesterol and lipoprotein metabolism.
- IL6 Interleukin 6 has several important roles in immunity.
- TGFB1 Transforming growth factor beta 1 controls cell proliferation and differentiation.
- MTHFR Methylenetetrahydrofolate reductase helps to process amino acids.
- ESR1 Estrogen receptor 1 has been a focus in breast, ovarian and endometrial cancers.
- AKT1 Encodes a signaling protein that phosphorylates other proteins to activate them.
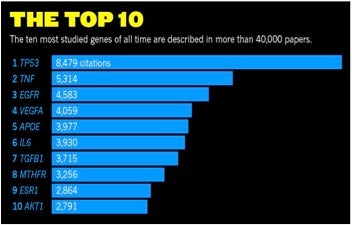 Adapted from Nature Vol 551, 2017; Source: Peter Kerpedjiev/NCBI-NLM
Adapted from Nature Vol 551, 2017; Source: Peter Kerpedjiev/NCBI-NLM
Topping the list is a gene called TP53, initial recognition of which goes back to publications in the early 1980s. The popularity of TP53 shouldn’t come as news to most biologists, as it is a tumor suppressor widely known as the ‘guardian of the genome’. The p53 gene is reported to be the most frequently mutated gene in human cancer: about 50% of all human cancers have lost p53 or express an inactive, mutant p53.
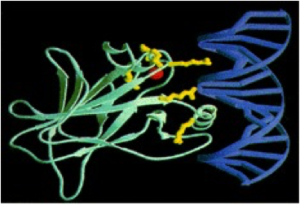 Core domain of p53 (green) bound to DNA (blue). Six most frequently mutated amino acids in yellow; red ball is zinc ion. Taken from Cho et al. Science, Vol 265, 1994
Core domain of p53 (green) bound to DNA (blue). Six most frequently mutated amino acids in yellow; red ball is zinc ion. Taken from Cho et al. Science, Vol 265, 1994
Given its preeminent position among all human genes studied, I had intended to provide a short synopsis of p53 functions identified to date. However, reading reviews quickly revealed that these functions are far too complex for me to adequately and succinctly summarize. Instead, I recommend consulting one such recent review by Haupt & Haupt, with the attention-grabbing title p53 at the start of the 21st century: lessons from elephants. According to these authors,
“[H>
ow p53 defends against DNA damage to preserve the genome and fight cancer is still being elucidated, despite more than 35 years of intense molecular and cellular study. Until very recently, the field predominantly focused on the role of p53 as a transcriptional regulator, particularly on its transactivation targets that drive arrest and apoptosis. The capacity of p53 to suppress tumors independent of key mediators of these processes has challenged accepted knowledge. The complexity of the p53 response continues to emerge along with new understanding of the contribution of p53 to transcriptional repression and also revisitation of the concept of p53 transactivation-independent function, which was first reported more than 20 years ago.”
Research into human cancer also brought scientists to TNF, the runner-up to TP53 as the most-referenced human gene of all time, with more than 5,300 citations in the NLM data. TNF encodes a protein—tumor necrosis factor—named in a 1975 publication because of its ability to kill cancer cells. But attempts to harness TNF’s anticancer action proved unsuccessful, as would-be therapeutic forms of the TNF protein were highly toxic when tested in people.
Like other popular genes, APOE is well studied because it’s central to cardiovascular disease, which is still one of the biggest unsolved health problems. First described in the mid-1970s as a transporter involved in clearing cholesterol from the blood, initial use of APOE protein as a lipid-lowering treatment was eventually supplanted by the statins. Now there is interest in an allele (i.e. genetic variant) of APOE, APOE4 that is associated with amyloid plaques and greatly increased risk of Alzheimer’s disease, about which I’ve blogged previously.
 Amyloid plaques. Taken from slideshare.net
Amyloid plaques. Taken from slideshare.net
Due to space limitations, I won’t comment on the remaining seven Top 10 genes, but will include this quote from the Nature news piece that comments on what influences a gene’s popularity, so to speak:
“[I>
t takes a certain confluence of biology, societal pressure, business opportunity and medical need for any gene to become more studied than any other. But once it has made it to the upper echelons, there’s a ‘level of conservatism’, says Gregory Radick, a science historian at the University of Leeds, UK, ‘with certain genes emerging as safe bets and then persisting until conditions change’. The question now is how conditions might change. What new discoveries might send a new gene up the chart—and knock today’s top genes off their pedestal?”
TriLink’s Connections to The Top 10 Genes
In a previous blog, I highlighted selected applications of TriLink’s products in publications during a given year. Extending this to the present Top 10 human genes, I searched Google Scholar for each of these genes cross-indexed with TriLink, and then selected a publication for each case. The results of each search can be perused later using the link embedded in each gene name, but for now here are snippets of each selected publication along with the TriLink product used and a link to the article.
- TP53 Immunization with p53 for rejecting established tumors; phosphorothioate CpG oligos; Daftarian et al.
- TNF Antisense inhibition of TNF synthesis; phosphorothioate oligos; Zhaowei et al.
- EGFR Aptamer-siRNA inhibition of EGFR; 2’-fluoro CTP; Giangrande & coworkers.
- VEGFA Aptamer for VEGF; 2’-O-methyl NTPs; Burmeister et al.
- APOE Modified mRNA delivery; 1methylpseudouridine-5′-triphosphate; Pardi et al.
- IL6 Biosensor for IL6 protein and DNA; biotinylated oligo; Wang et al.
- TGFB1 RNA-seq of leukemia cells; Cy3 and Cy5 labeled random 9-mers; Wilhelm et al.
- MTHFR Genotyping MTHFR mutants in blood; Molecular Beacon probes; Shi et al.
- 9. ESR1 SNP genotyping ESR1 associated with breast cancer; custom synthesized oligos as probes; Gold et al.
- AKT1 Antisense inhibition of AKT1; phosphorothioate oligos; Yoon et al.
Concluding Comments
When researching and writing this blog, several thoughts crossed my mind. Firstly, investigations of genes related to cancer in one way or another comprise a significant portion of The Top 10 literature. Given the prevalence and seriousness of cancer worldwide, this is not surprising, and perhaps expected.
Secondly, the news article in Nature that I cited in the introduction states that out of the 20,000 or so protein-coding genes in the human genome, only 100, which is only 0.5% of the 20,000 protein-coding genes, account for more than 25% of all the papers tagged in the NCBI/NLM PubMed database. The news item added that thousands of genes go unstudied in any given year, and quoted Helen Anne Curry, a science historian at the University of Cambridge, UK as saying ‘It’s revealing how much we don’t know about because we just don’t bother to research it.’ Although this reminds me a bit of the currently popular, self-evident saying “we don’t know about what we don’t know”, the implication is that we don’t know much about an extensive portion of the human protein-coding genome.
Thirdly, from a company perspective, it was evident that TriLink’s long-time expertise in modified nucleic acids has enabled, over the years, many investigations of all types related to these Top 10 genes, which is good thing.
Finally, from a personal perspective, it was gratifying to find that my early development of automated synthesis of phosphorothioate oligos evolved to provide ready access to this remarkably unique and useful class of modified nucleic acids pioneered by my long-time friend Fritz Eckstein.
As usual, your comments are welcomed.






