- Simulated Sequencing Takes Virtual Reality Way Beyond Games
- In Silico Simulations Suggest Possible 99.99% Accuracy for Graphene Nanopores
- minION Nanopore Sequencer is Sent to the International Space Station
Prelude
 Taken from oculusvr.com
Taken from oculusvr.com
This blog is mostly about an international team of researchers who are using Virtual Reality (VR)—in the form of computational modeling—to simulate a new approach to DNA sequencing using nanopores made out of graphene. While VR is a hot trend in all sorts of so-called immersion media, such as those offered by Oculus (that was acquired by Facebook for $2 billion in 2014), computation-based VR has been used by scientists for simulating molecular interactions for a relatively long time. However, extending molecular simulations to complex (aka many-atom) systems like nanopores and DNA has had to wait for bigger, faster, cheaper computing.
In this blog, I’ll also discuss the recent launch of a commercially available nanopore sequencer for the first ever DNA sequencing in space using a self-landing rocket operated by Space X (co-founded by uber-famous multi-billionaire entrepreneur Elon Musk). It’s hard for me to even imagine what seemingly incongruent mix of topics could be more intriguing than these. As the now trendy saying goes, you can’t make this stuff up. But I digress…
 Lift off! Taken from am1070theanswer.com - Self-landing! Taken from indianexpress.com
Lift off! Taken from am1070theanswer.com - Self-landing! Taken from indianexpress.com
Nanopore Sequencing
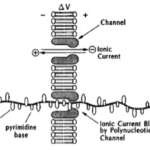 From an earlier blog you’ll know that I’m a huge fan of tiny nanopores for sequencing, which is a 20+ year old concept, as depicted below from a seminal patent wherein DNA was envisaged as moving through a pore-in-lipid bilayer leading to base-dependent transient blockage of ionic current from which sequence is determined.
From an earlier blog you’ll know that I’m a huge fan of tiny nanopores for sequencing, which is a 20+ year old concept, as depicted below from a seminal patent wherein DNA was envisaged as moving through a pore-in-lipid bilayer leading to base-dependent transient blockage of ionic current from which sequence is determined.
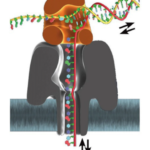
After two decades, this prophetic concept of nanopore sequencing has recently been realized, and commercialized by Oxford Nanopore Technologies (ONT) using “bionanopore” technology. Comparing the images above and below, you’ll see that bionanopores are, in many respects, quite similar to the first described nanopores, wherein a pore-forming protein, α-hemolysin (gray), is embedded in a lipid bilayer (blue). On the other hand, there is an attached DNA-processive enzyme, 29 DNA polymerase (brown), that feeds in the single strand of DNA for sequencing; details may be found elsewhere.
An alternative strategy for nanopore sequencing is to replace this type of bionanopore (composed of biological macromolecules) with a pore constructed of non-biological materials, notably silicon-based semiconductors that enable electrical signal generation and data processing. This would-be evolution of nanopore sequencing from biological constructs to various types of solid-state materials can be read about elsewhere.
Whither Goest Graphene?
It seems the next step in the progression of nanopore technology is those made of graphene—the trivial name for a very special form of carbon that was long known but exceedingly difficult to make. In fact, the process is so difficult that Andre Geim and Kostya Novoselov at The University of Manchester were awarded the 2010 Nobel Prize in Physics for their work enabling the production and characterization of graphene.

Graphene is a two-dimensional array or “sheet” of carbon atoms that is usually depicted by the ball-and-stick model (pictured at the left below) as a one-atom-thick sheet of otherwise infinite dimensions. Since nothing is infinite in the real world, sheets of graphene have edges to which hydrogen is bonded, but for simplicity is ignored. This carbon-carbon bonding with carbon-hydrogen edges is akin to that in polycyclic aromatic hydrocarbons familiar to readers who are chemists.

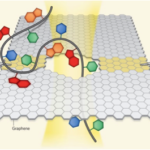
Because of graphene’s unique electrical properties and single-atom-thin structure, the basic idea is that a nanometer-size hole in graphene might be made—somehow—to allow DNA and ions to pass through and thus generate electrical signals—somehow—that are accurately deciphered—somehow—into DNA sequence. Oh, and let’s not forget that this sequence information must differentiate—somehow—3’->5’ from 5’->3’ directional pass through. All these “somehows” are meant to indicate that it’s far easier to imagine the concept of graphene nanopore sequencing, as fancifully shown below, than to actually do it.
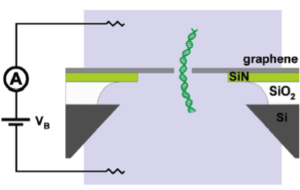
The most daunting practical problem deals with how to “drill” tiny holes in graphene. One approach has been to use controlled electron-beam exposure in a transmission electron microscope. Initial demonstration of this approach was published in 2010 by Merchant et al. in Nano Letters in a paper titled DNA Translocation through Graphene Nanopores, from which the schematic left is taken.
In this device, a few-atoms-layer piece of graphene (1-5 nm thick) having an ~10 nm hole is suspended over a 1 μm diameter hole in a 40 nm thick silicon nitride (SiN) membrane suspended over an ~50 × 50 μm2 aperture in a silicon chip coated with a 5 μm silicon oxide (SiO2) layer in such a way that a bias voltage (VB) is applied between the reservoirs to drive DNA through the nanopore. Although DNA could be detected, the graphene pore size was too big to allow sequence detection.
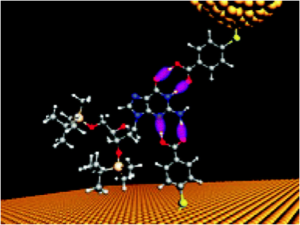
Similar studies by Schneider et al. were also reported in Nano Letters in 2010, which appears to be a watershed year for this journal inasmuch as another noteworthy nano-detection scheme for DNA was described therein by Chang et al.—but with an important new feature. Namely, using gold electrodes (in yellow, below) separated by only 2 nm and conjugated to dC, a derivative of dG (blue balls) apparently was able to H-bond (magenta) to dC—based on dC-dG complementarity and detected as electron tunneling signals. This transient, base pair-specific H-bonding is what has now been further investigated by others albeit in the following form of Virtual Reality.
Virtual Reality Nanopore Sequencing
In contrast to the above “real” experiments, others have simulated reality using mathematical calculations based on theoretical chemistry, which is Virtual Reality that has physical significance well beyond simply playing games. Mathematical modeling or computation simulations are phrases generally used to describe these so-called in silico “experiments” that serve as indications of what could be done, in theory, if this Virtual Reality is actually translatable to the real world. But I digress…
An international team of investigators in the U.S., Germany, and Netherlands has recently reported studies titled Nucleobase-functionalized graphene nanoribbons for accurate high-speed DNA sequencing. Although this article is a dreaded “pay-to-read” article, there is a brief news piece about it at the website for the U.S. National Institute of Standards and Technology (NIST) where some of this work was conducted.
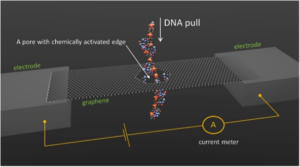
As is evident from the schematic shown below, these investigators borrowed from the aforementioned types of publications to imagine a graphene nanopore having its internal edges functionalized with nucleobase moieties that could potentially H-bond with DNA bases in a sequence specific manner—à la Chang et al. Under appropriate conditions, this could provide the basis for sequencing via measurement of induced current fluctuations.
More specifically, they imagined a sheet (aka ribbon) of graphene 4.5 x 5.5 nm with several nucleobase moieties attached to a 2.5 nm nanopore. In animated simulations (which are linked at the NIST website), you can watch how this sensing device would perform at room temperature in water with attached cytosine H-bonding to detect G in DNA.
When you watch this simulation, you’ll immediately notice how “wiggly” DNA is due to random motions of its constituent groups and atoms. You’ll also see detection of each translocating (i.e. passing G) as an increasing signal being recorded in real-time. While time as a parameter in this simulation is real, the simulation itself is not real, but rather virtual reality based on state-of-the art theoretical calculations by a computer.
With that caveat in mind, the performance was said to be 90% accurate (due to missed bases rather than wrongly detecting a base) at a rate of 66 million bases per seconds, which to me is mindboggling ultra-fast. Moreover, if this device could be fabricated as four sequentially-located graphene pores each functionalized with either C, G, A, or T, the researchers estimate that “proofreading” would increase accuracy to 99.99%, as required for sequencing the human genome.
Virtual reality is, well, not reality. And sometimes dreams and reality go in opposite directions. However, if indeed the above imaginary device and simulations were to become reality—nanopore sequencing would indeed be advanced dramatically from today’s performance.
ONT’s minION Sequencer in Space
In transitioning back to reality, it’s almost unbelievable to me that ONT’s minION nanopore sequencer—which I’ve blogged about before—was sent to the International Space Station (ISS) in April 2016 to carry out the first ever sequencing of DNA in space. If that wasn’t enough “buzz”, then the fact that this was achieved by uber-famous Elon Musk’s Space X company made it way more so, along with much ado in successfully landing the rocket’s first-stage on a relatively tiny platform in the ocean. This is all amazing stuff. And to think that not so long ago, we were thinking how great it would be if self-landing rockets were really possible, not just a fun concept in video games and sci-fi movies! Maybe virtual reality isn’t that far from becoming reality, but I digress….
The first aim of putting the minION nanopore into space is to demonstrate the feasibility of nanopore sequencing in microgravity. That being done would then allow use of the minION to rapidly sequence astronaut samples in the ISS to diagnose, for example, an infectious disease or other health issue.
Interested readers can peruse elsewhere much more about this historic milestone, as well as watch and listen to a short (but very exciting) video titled Space Station Live: Big DNA Science in a Small Package. The video was posted on Twitter on July 21st and features the minION device (aka The Biomolecular Sequencer).

I look forward to learning the results of the minIONs research in space. I hope that it’s “mission accomplished!” and of great use in years to come. BTW, if you’re a “space buff” like me, you can watch and listen to ISS-Mission Control live streaming 24-7 at this website.
As usual, your thoughts about this blog are welcomed as comments.







