- Adjuvants Can Lower Vaccine Doses
- Prof. Philip J. Santangelo Has Demonstrated a Novel “Tethered Adjuvant” Approach for mRNA Vaccines
- Tethered Adjuvants are Targeted to Candidate mRNA Vaccines Using SoluLINK® Bioconjugation Chemistry
Backstory
The current COVID-19 pandemic has led to an unprecedented amount of attention and activity on the “warp speed” development of a safe and effective vaccine. Out of over more than 100 various types of vaccines under development for COVID-19 reported by Le et al., mRNA vaccine technology is in the spotlight, as discussed in the April 28th Zone blog. Importantly, Le et al. note that for some vaccine platforms, “adjuvants could enhance immunogenicity and make lower doses viable, thereby enabling vaccination of more people without compromising protection.” So far, they add, “at least 10 developers have indicated plans to develop adjuvanted vaccines against COVID-19.”

Given this potentially critical aspect of adjuvanted vaccines and the spotlight on mRNA vaccine technology, the Zone has researched the literature for adjuvanted mRNA vaccines. Among the articles found, a publication by Loomis et al. titled In vitro transcribed mRNA vaccines with programmable stimulation clearly stood out. This work, led by Prof. Philip J. Santangelo in the Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, describes a fundamentally different and completely novel way to employ adjuvants for mRNA vaccines. Before presenting a synopsis of this development, I will outline the scientific rationale of the design strategy for the approach, as well as provide an introduction on vaccine adjuvants and their function.
Design Strategy
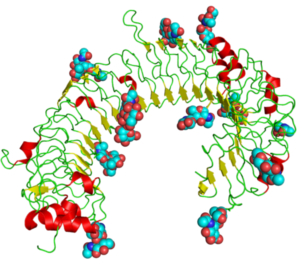 The horseshoe structure of TLR3, showing attached sugars (spheres) and internal structures (wires, arrows, and helixes). Taken from commons.wikimedia.org and free to use.
The horseshoe structure of TLR3, showing attached sugars (spheres) and internal structures (wires, arrows, and helixes). Taken from commons.wikimedia.org and free to use.
Loomis et al. state that in vitro translated (IVT) mRNA delivered to host cells is translated by the cellular activated pattern recognition receptors (PRRs) that have evolved as antiviral RNA sensors to detect single- and double-stranded (ss and ds) transcripts. As reviewed elsewhere, PRRs can be classified according to their primary location within the cell: Toll-like receptors (TLRs) 3, 7 (shown here), and 8 all bind ssRNA and dsRNA in the endosomal compartment; while cytoplasmic detection largely relies on RIG-I-like receptors (RLRs) and Nod-like receptors (NLRs). TLR signaling causes translocation of transcription factors (i.e., NF-kappaB, IRF7, IRF3) to the nucleus, ultimately releasing Type 1 interferons (IFNs) and pro-inflammatory cytokines. These secreted IFNs and cytokines act in an autocrine and paracrine fashion to sensitize nearby cells to nucleic acids.
As depicted here, TLR expressing antigen-presenting cells (APCs) such as dendritic cells (DCs) and macrophages are not only the orchestrators of the innate immune response, but they are also responsible for linking the innate and adaptive immune responses upon migration to draining lymph nodes and activation of T and B cells. The type of antigen specific effector and memory responses that are generated largely depend on the cytokine profiles released by cells during the innate response. For example, IL-12 and IFNγ promote TH1 and cytotoxic T lymphocyte (CTL) pathways, while IL-4 and IL-6 promote humoral responses.
Unmodified “naked” IVT mRNA has been shown to elicit robust type I interferon responses, which can activate protein kinase R (PKR), 2’-5’-oligoadenylate synthetase, and caspase-1 mediated cell death, resulting in reduced antigenic protein levels due to protein translational inhibition and RNA decay. High IFN responses have also been linked to weak cell-mediated immunity.
The innate immune responses elicited for an IVT mRNA vaccine influence the yield of antigenic protein, as well as the nature of specific memory immune responses. This has led to significant efforts in studying the intrinsic immunogenicity of unmodified vs. modified IVT mRNA and the use of adjuvants to modulate immunogenicity. Substitution of uridine with the modified bases pseudouridine (Ψ) or N1-methylpseudouridine (M1 Ψ) in IVT mRNA has been shown to reduce innate immune activation and to increase transgenic protein synthesis (e.g. Pardi et al.). However, reduced innate immune responses to modified IVT mRNAs often require the use of specific adjuvants to grant supplemental immunogenicity by controlling innate cytokine release (e.g. Verbeke et al.). The potential benefits of utilizing adjuvants include dose and frequency reduction, vaccine response broadening, rapid response, and overcoming immune senescence.
In immunology, an adjuvant is a substance that potentiates and/or modulates the immune responses to an antigen to improve them. The word "adjuvant" comes from the Latin word adiuvare, meaning to help or aid. "An immunologic adjuvant is defined as any substance that acts to accelerate, prolong, or enhance antigen-specific immune responses when used in combination with specific vaccine antigens." In the early days of vaccine manufacturing, significant variations in the efficacy of different batches of the same vaccine were correctly assumed to be caused by contamination of the reaction vessels. However, it was soon found that more scrupulous cleaning actually seemed to reduce the effectiveness of the vaccines, and that some contaminants actually enhanced the immune response. There are many known adjuvants in widespread use, including aluminum salts, oils, and virosomes. Source: wikipedia.org

he main challenge in the successful design of IVT mRNA vaccines is the development of a versatile platform able to grant high antigenic protein expression, while inducing controlled and specific robust memory immune responses.”
To address this critical challenge, Loomis et al. envisaged an IVT mRNA-based vaccine platform that enables modular tethering of small molecule adjuvants for programmed stimulation of innate immunity. For proof-of-concept, an IVT mRNA encoding for the well-established cytoplasmic ovalbumin antigen (OVA) was selected as a model vaccine.
OVA Antigens. Ovalbumin (OVA) is a key reference protein for vaccination experiments. Ovalbumin, the major protein constituent of chicken egg whites, is a glycoprotein that is sufficiently large and complex to be mildly immunogenic. Consequently, it is widely used as an antigen for immunization research. Source: wikipedia.org
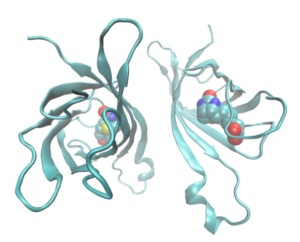
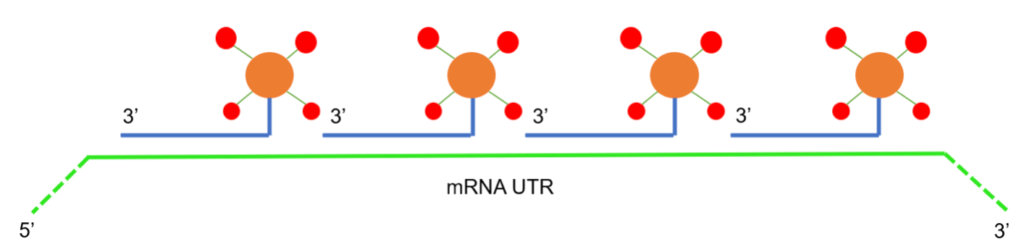
To tether small molecule agonists to IVT mRNA, Loomis et al. modified a method for designing multiply labeled tetravalent RNA imaging probes (MTRIPs), earlier pioneered by the Santangelo lab, as detailed elsewhere. MTRIPs are comprised of four RNA-targeting 2’-O-methyl-RNA/DNA chimeric oligonucleotides (oligos) assembled on a tetramer core of NeutrAvidin, which is a deglycosylated version of avidin, depicted here as a dimer for better visualization of its biotin ligand. MTRIPs have been used to study endogenous and exogenous RNA dynamics via fluorescence microscopy without hindering translation or affecting stability (e.g. Kirschman et al.).
In contrast to these original MTRIPs, monovalent MTRIPs (mMTRIPs) are synthesized by covalently linking single RNA-targeting oligos to free amines on a NeutrAvidin core. Four different oligos are selected, each targeting a different region in the 3’ untranslated region (UTR) of the IVT mRNA. The four available biotin binding sites on the NeutrAvidin core are then utilized to conjugate biotinylated adjuvants, thus offering a flexible platform for the delivery of small molecule agonists.
The proof-of-concept publication by Loomis et al. is divided into four parts, as outlined in the next sections. First, they abrogated the intrinsic innate response profile of OVA mRNA through M1Ψ base incorporation, which also led to enhanced production of the transgene protein in vitro. Then, they synthesized adjuvant-tethered mTRIPs targeting the OVA mRNA. Next, they used a transfected mouse macrophage cell line (RAW264.7) to demonstrate that tethering TLR agonists to mRNA can activate TLR2 or TLR7 in vitro. Lastly, they showed that increased cellular and humoral immune responses following murine intramuscular (i.m.) injection of TLR7 agonists tethered to naked IVT mRNA when compared to a cocktail formulation.
Attenuation of Innate Response Produced by mRNA Transfection
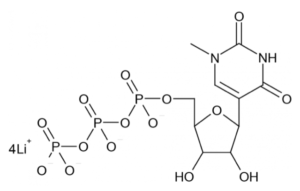
Substitution of uridine with the modified bases Ψ or M1 Ψ in IVT mRNA can reduce innate immune activation and increase transgenic protein synthesis. Loomis et al. therefore synthesized three OVA expressing IVT mRNAs identical in sequence, either incorporating unmodified bases, or substituting Ψ or M1 Ψ (depicted here) for uridine. Innate immune activation and protein expression were studied in vitro in RAW 264.7 macrophage cells transfected using Lipofectamine 2000 (L2K), which is a commonly used transfection agent despite not being biologically inert and inducing a moderate immune activation.
Total RNA was isolated and assayed for genes associated with immune responses to exogenous RNA, such as viral RNA. As expected, IVT mRNA composed of unmodified bases led to the highest upregulation of IFN signaling and stimulated genes over L2K-only controls, while IVT mRNA incorporating M1 Ψ was less immune-stimulatory than IVT mRNA incorporating Ψ. The ability to achieve a minimal level of mRNA-induced innate immune activation is advantageous in addressing the effect of specific TLR agonists (i.e. adjuvants) and modulating activation of innate receptors, while still retaining enhanced protein expression. Therefore, M1 Ψ -IVT mRNA was utilized for all subsequent studies.
Synthesis of mMTRIPs and In Vitro Characterization of Adjuvant-Tethered IVT mRNA
The mMTRIPs were synthesized by designing the following four 2’-O-methyl RNA/DNA chimeric oligos complementary to the 3’ UTR of the IVT mRNA for OVA, the model antigen:
5’ TTTTTTTGCAAGCCCCGCAGAAGG 3’
5’ TTTATTTAGAGAAGAAGGGCAUGG 3’
5’ TTTTTTACCAAGAGGUACAGGUGC 3’
5’ TTTTTTTCUACUCAGGCUUUAUUC 3’
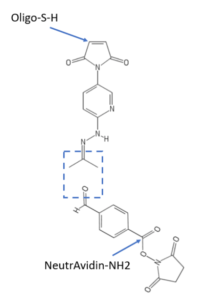
The boldface indicates 2’-O-methyl RNA moieties in the oligos, each of which has a 5’ C6-disulfide moiety for conjugation to NeutrAvidin using SoluLINK® bioconjugation chemistry. Briefly, the 5’ disulfide was reduced by incubation with tris(2-carboxyethyl)phosphine (TCEP) and the resultant 5’ thiol oligos were filtered to remove unreacted TCEP. As depicted here, the 5’ thiol oligos were individually modified with a hydrazinonicotinamide (HyNic) functional group by overnight incubation with maleimide HyNic. Each HyNic-modified oligo was then buffer exchanged by centrifugal filtration. Separately, NeutrAvidin was labeled with succinimidyl-4-formylbenzamide (S4-FB) and buffer exchanged by centrifugation. To conjugate S4-FB-functionalized NeutrAvidin to each HyNic-functionalized oligo, the reactants were incubated at a 1:2 molar ratio in SoluLINK TurboLINK® Buffer. Unconjugated oligos were removed by centrifugal filtration and the final NeutrAvidin concentration was determined by the bicinchoninic acid assay against a standard curve. Oligo content was measured by UV absorbance at 260 nm. The reaction was optimized to yield approximately a 1:1 molar ratio of NeutrAvidin : oligo for each mRNA targeting sequence.
SoluLINK® bioconjugation technology is available from Vector Laboratories, which is part of Maravai LifeSciences, as is TriLink BioTechnologies. Readers interested in any type of bioconjugation are encouraged to consult the white paper on bioconjugation using SoluLINK® reagents available at this link. The white paper provides a concise introductory overview, and also discusses advantages of SoluLINK® compared to other methods used to prepare covalent conjugates involving azide-alkyne “click” chemistry or maleimide-thiol coupling.
As depicted below, commercially available biotinylated small molecule agonists (red) were bound to the NeutrAvidin (orange)-oligo (blue) conjugates, each of which has 4 sites for binding to biotin (not shown). The four resulting mMTRIPs were purified via filtration and then hybridized to the UTR of the IVT mRNA of OVA (green). To determine the efficiency of small molecule tethering, a biotinylated fluorescent dye was used as a measurable surrogate. Following purification, the overall fluorescence and 260 nm absorbance of the sample were measured and compared to a standard curve of known molar concentration of pure dye. It was determined that, on average, 10 fluorescent molecules were bound to an IVT mRNA, which indicates that the IVT mRNA for OVA was labeled with approximately 2.5 mMTRIPS on average, and a maximum of four mTRIPs.
The mMTRIPs could potentially alter the secondary structure of the IVT mRNA, leading to PRR activation. Consequently, unmodified or M1 Ψ -modified IVT mRNA were delivered to RAW264.7 cells with or without tethered mMTRIPs, and TLR7 activation was quantified, as detailed in Loomis et al.’s supporting information, freely available at this link. As expected, unmodified IVT mRNA upregulated TLR7 with respect to the L2K-only control condition, while the M1 Ψ IVT mRNA did not. However, no statistically significant differences in TLR7 activation were observed between unlabeled and mMTRIP-labeled mRNA, indicating that mMTRIP hybridization does not affect TLR7 activation. To evaluate potential immunogenicity of the 2’-O-methyl chimeric oligos, cytokines were quantified (see supporting information) in Vero cells transfected with tethered or untethered mTRIPs. No significant difference was observed. These data support the assumption that innate immune activation observed upon delivery of adjuvant-tethered mRNA would be solely reflective of the agonist payload.
Tethering Adjuvant to IVT mRNA Induces an Acute Innate Response In Vivo
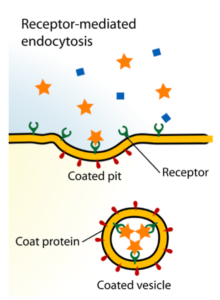
Next, Loomis et al. investigated whether agonist-tethering to unformulated naked IVT mRNA would lead to stronger innate immune responses than a mixed-preparation “cocktail” following i.m. injection in a murine model system. Since APCs, which should ideally be targeted and activated, have a strong proclivity for naked mRNA uptake upon i.m. injection, according to Loomis et al., no carriers or formulations were utilized for delivery. They reasoned that upon i.m. injection, a cocktail preparation would elicit an effectively weaker immune response, because the small molecule adjuvants would readily diffuse away from the injection site, given their negligible molecular weights. Contrarily, tethering the small molecule adjuvants to naked IVT mRNA, the uptake of which involves clathrin-mediated endocytosis (depicted here), would promote adjuvant retention at the site of injection and increase interactions with endosomal TLRs upon internalization, eliciting a stronger and longer-lasting localized effect.
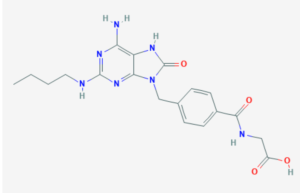
To assess this, M1 Ψ IVT mRNA with tethered CL264 adjuvant (shown here) or as a cocktail was injected i.m. (five mice per experimental condition). M1 Ψ mRNA and CL264 were also injected independently as controls. Five hours post-injection, muscles were isolated and analyzed for local antiviral gene expression by qRT-PCR (see supporting information). It was found that injection of naked M1 Ψ IVT mRNA, characterized by an intrinsically low immunogenicity, can be delivered i.m. with a tethered payload of CL264 adjuvant in vivo, leading to an enhanced local innate immune response at the injection site.
Adaptive Responses to Adjuvant-Tethered IVT mRNA In Vivo
Next, the researchers analyzed the humoral immune response in the serum extracted from the same animals by measuring the concentration of anti-OVA specific IgG in sera using an enzyme-linked immunosorbent assay (ELISA). All 5 animals that received tethered CL264-mRNA formulations contained detectable levels of anti-OVA IgG, with an average concentration 8-fold higher than those treated with a cocktail preparation. The elicited OVA-specific adaptive responses suggest that tethering adjuvants to IVT mRNA strongly increases mRNA immunogenicity and adjuvant activity in vivo.
Conclusions
Loomis et al. described a novel approach using tethered small molecule adjuvants to control IVT mRNA-induced innate immune responses, which then drove the development of robust cell mediated and humoral immunity in a mouse model system. Delivery vehicles confer IVT mRNA protection from nucleases and allow for cell-type specific delivery, but they are not immunologically inert. Moreover, size, shape, and charge of delivery vehicles have been found to affect both cell type uptake and immunological stimulation. The present approach does not rely on the incorporation of delivery vehicles for immune stimulation per se, but both methods are potentially complementary for customized vaccine design.
Tethering small molecule adjuvants to the much larger mRNA likely decreases its diffusion from the injection area, while raising its local concentration at the injection site and promoting interactions with PRRs. According to Loomis et al. tethered adjuvants “potentially may represent a strategy to lower the systemic toxicity associated with numerous adjuvants.”
Finally, Loomis et al. “believe that, by rationally choosing adjuvant-mRNA combinations, this modular platform can help standardize vaccine studies and allow for specific and directed immune responses in streamlining the design and improvement of mRNA-based vaccines.”
Final Comments
The Zone fully agrees with these conclusions by Loomis et al., and will be diligently monitoring the literature for any follow-up publications about this novel adjuvant tethering approach for vaccine development—especially for COVID-19, as mentioned in the backstory for this blog.
Your comments are welcomed, as usual.








