Loading...
7 view(s)
- There are Now Millions of DNA-Related Publications
- Some of the Top 5 Cited Papers on DNA Will Surprise You
- You Probably Won’t Guess Top 5 Most Frequently Cited

Deciding what to post here in recognition of DNA Day 2017 was just as challenging as it has been in past years, primarily because there’s so many different perspectives from which to choose. After much mulling, and several abandoned approaches, I settled on featuring DNA publications that have received the most citations, as an objective metric—not just my subjective opinions about topics I think are significant or otherwise interesting.
Before getting to the numbers of DNA-related papers and some of the most cited papers, here’s a quick recap of what was posted here in the past, starting with the inaugural blog four years ago:
2013—60
th Anniversary of the Discovery of DNA’s Double Helix Structure
2014—My Top 3 “Likes” for DNA Day
2015—Celebrating Click Chemistry in Honor of DNA Day
2016—DNA Dreams Do Come True!
Explosive Growth of DNA Publications
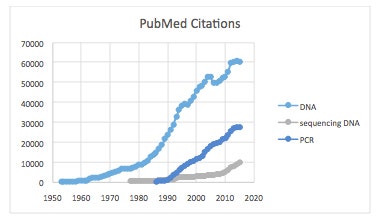
Regular readers of my blogs will know that I frequently use the NIH PubMed database of scientific articles to find publications by searching keywords, phrases, or authors. A convenient feature of these searches is providing “results per year” that can be exported into Excel for various purposes. Some preliminary searches indicated that DNA-related articles can be indexed by either DNA or PCR, or cloning, or other terms among which sequencing was notable. The majority, however, were indexed as either DNA or PCR, which together gave nearly 1.7 million items—an astounding number. This number is even much greater since PubMed excludes some important chemistry journals, as well as patents.
Diving deeper into these numbers, I thought it helpful to look at the publication volumes and rates for DNA, sequencing DNA, and PCR through 2015 starting from 1953, 1977, and 1986, respectively. These respective dates correspond to seminar publications by
Watson & Crick,
Maxam & Gilbert, and
Mullis & coworkers. The results shown in the following graph attest to my often stated “power of PCR” as premier method in nucleic acid research, which we’ll see again below in another numerical context.
 Top 5 Cited Papers
Top 5 Cited Papers
During my perusal of the above literature in PubMed generally related to DNA, I thought it would be interesting to find, and share here, which specific papers have the distinction of being most frequently cited. Citations are not available in PubMed, but are compiled in Google Scholar, which led me to these Top 5 that are listed from first to fifth.
- DNA sequencing with chain-terminating inhibitors
Frederick Sanger, the eponymous father of the “Sanger sequencing” method published in 1977, received the 1980 Nobel Prize in chemistry for this contribution. He also received the 1958 Nobel Prize in chemistry for sequencing insulin, and is the only person to win two Nobel Prizes in chemistry. Uber-famous DNA expert Craig Venter is
quoted as saying that ‘Fred Sanger was one of the most important scientists of the 20th century,’ [who>
‘twice changed the direction of the scientific world.’
- Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method
 Kenneth J. Livak, PhD
Kenneth J. Livak, PhD
Taken from archive.sciencewatch.com
The most commonly used method to analyze data from
real-time, quantitative PCR (RT-qPCR) experiments is relative quantification, which relates the PCR signal of the transcript of interest to that of a control sample such as an
untreated control. The derivation, assumptions, and applications of this method were published in 2001 by
Livak & Schmittgen. I overlapped with Ken Livak at Applied Biosystems, which pioneered commercilaization of RT-qPCR reagents and instrumentation at the time. He is currently Senior Scientific Fellow at Fluidigm Corp.
 Sir Edwin M. Southern Taken from ogt.co.uk
Sir Edwin M. Southern Taken from ogt.co.uk
3.
Detection of specific sequences among DNA fragments separated by gel electrophoresis
Sir Edwin Mellor Southern, FRS, the eponymous father of “Southern blotting” DNA fragments from agarose gels to cellulose nitrate filters published in 1975, is a
Lasker Award-winning molecular biologist, Emeritus Professor of Biochemistry at the University of Oxford and a fellow of Trinity College. He is also Founder and Chief Scientific Advisor of Oxford Gene Technology.
-
 Prof. Bert Vogelstein, MD
Prof. Bert Vogelstein, MD
Taken from hhmi.org
This paper by Feinberg & Vogelstein published in 1983 describes how to conveniently radiolabel DNA restriction endonuclease fragments to high specific activity using the large fragment of DNA polymerase I and
random oligonucleotides as primers. These "oligolabeled" DNA fragments serve as efficient probes in filter hybridization experiments. His group pioneered the idea that somatic mutations represent uniquely specific biomarkers for cancer patients, leading to the first FDA-approved DNA mutation-based screening tests, and now
“liquid biopsies” that evaluate blood samples to obtain information about underlying tumors and their responses to therapy (an area that I’ve touted in previous blogs). A technique for conveniently radiolabeling DNA restriction endonuclease fragments to high specific activity is described. DNA fragments are purified from agarose gels directly by ethanol precipitation and are then denatured and labeled with the large fragment of DNA polymerase I, using random oligonucleotides as primers. Over 70% of the precursor triphosphate is routinely incorporated into complementary DNA, and specific activities of over 10
9 dpm/μg of DNA can be obtained using relatively small amounts of precursor. These “oligolabeled” DNA fragments serve as efficient probes in filter hybridization experiments.
-
 Kary B. Mullis, PHD
Kary B. Mullis, PHD
Taken from TED.com
In 1988,
Kary B. Mullis and coworkers (then at Cetus Corp.) published in venerable
Science a method using oligonucleotide primers and thermostable DNA polymerase from
Thermus aquaticus to amplify genomic DNA segments up to 2000 base pairs to detect a target DNA molecule present only once in a sample of 10
5 cells. Since that time, polymerase chain reaction (PCR)-related technology has evolved to now routinely enable a variety of single-cell analyses of DNA or RNA. Dr. Mullis received the 1993 Nobel Prize in chemistry for his 1983 invention of PCR, which his
website says
‘is hailed as one of the monumental scientific techniques of the twentieth century.’
Top 5 Papers by Citation Frequency
While writing the above section, it occurred to me that ranking these five publications by total number of citations-to-date in Google Scholar doesn’t account for differences in the number of years between the year of publication and now. I did the math to calculate the average citation frequency per year, and here’s the totally surprising—to me—result: relative gene expression methodology published by
Livak & Schmittgen is by far the most frequently cited of the Top 5, according to this way of ranking:
- 2001, relative gene expression, Cited by 69560 = 4,637 avg. citations per year
- 1977, Sanger sequencing, Cited by 32662 = 1,701
- 1975, Southern blotting, Cited by 21201 = 796
- 1988, PCR, Cited by 18785 = 671
- 1983, oligolabeled DNA, Cited by 21200 = 642
I should point out that, as transformative methods such as these gradually become widely recognized as “standard procedures,” researchers tend to feel it unnecessary to include a reference to the orignal publication. Consequenly, citation frequency decreases with time even though cummulative usage increases. In other words, 25 years from now average citations per year for relative gene expression will have likely decreased, and be surpassed by a new “method of the decade,” so to speak.
Prediction for the Future
This line of reasoning leads me to close with some speculation about what DNA-related technique might emerge as the next “method of the decade” that tops the above ranking by citation frequency.
My guess is that it will be
Multiplex genome engineering using CRISPR/Cas systems by Zhang & coworkers that has been cited by
4145 at the time I’m writing this piece, only four years from its publication in venerable
Science in 2013. Some of my
blogs have already commented on various aspects of CRISPR/Cas9, which is among
genome editing tools offered by TriLink.
As usual, your comments are welcomed.
 Deciding what to post here in recognition of DNA Day 2017 was just as challenging as it has been in past years, primarily because there’s so many different perspectives from which to choose. After much mulling, and several abandoned approaches, I settled on featuring DNA publications that have received the most citations, as an objective metric—not just my subjective opinions about topics I think are significant or otherwise interesting.
Before getting to the numbers of DNA-related papers and some of the most cited papers, here’s a quick recap of what was posted here in the past, starting with the inaugural blog four years ago:
2013—60th Anniversary of the Discovery of DNA’s Double Helix Structure
2014—My Top 3 “Likes” for DNA Day
2015—Celebrating Click Chemistry in Honor of DNA Day
2016—DNA Dreams Do Come True!
Explosive Growth of DNA Publications
Regular readers of my blogs will know that I frequently use the NIH PubMed database of scientific articles to find publications by searching keywords, phrases, or authors. A convenient feature of these searches is providing “results per year” that can be exported into Excel for various purposes. Some preliminary searches indicated that DNA-related articles can be indexed by either DNA or PCR, or cloning, or other terms among which sequencing was notable. The majority, however, were indexed as either DNA or PCR, which together gave nearly 1.7 million items—an astounding number. This number is even much greater since PubMed excludes some important chemistry journals, as well as patents.
Diving deeper into these numbers, I thought it helpful to look at the publication volumes and rates for DNA, sequencing DNA, and PCR through 2015 starting from 1953, 1977, and 1986, respectively. These respective dates correspond to seminar publications by Watson & Crick, Maxam & Gilbert, and Mullis & coworkers. The results shown in the following graph attest to my often stated “power of PCR” as premier method in nucleic acid research, which we’ll see again below in another numerical context.
Deciding what to post here in recognition of DNA Day 2017 was just as challenging as it has been in past years, primarily because there’s so many different perspectives from which to choose. After much mulling, and several abandoned approaches, I settled on featuring DNA publications that have received the most citations, as an objective metric—not just my subjective opinions about topics I think are significant or otherwise interesting.
Before getting to the numbers of DNA-related papers and some of the most cited papers, here’s a quick recap of what was posted here in the past, starting with the inaugural blog four years ago:
2013—60th Anniversary of the Discovery of DNA’s Double Helix Structure
2014—My Top 3 “Likes” for DNA Day
2015—Celebrating Click Chemistry in Honor of DNA Day
2016—DNA Dreams Do Come True!
Explosive Growth of DNA Publications
Regular readers of my blogs will know that I frequently use the NIH PubMed database of scientific articles to find publications by searching keywords, phrases, or authors. A convenient feature of these searches is providing “results per year” that can be exported into Excel for various purposes. Some preliminary searches indicated that DNA-related articles can be indexed by either DNA or PCR, or cloning, or other terms among which sequencing was notable. The majority, however, were indexed as either DNA or PCR, which together gave nearly 1.7 million items—an astounding number. This number is even much greater since PubMed excludes some important chemistry journals, as well as patents.
Diving deeper into these numbers, I thought it helpful to look at the publication volumes and rates for DNA, sequencing DNA, and PCR through 2015 starting from 1953, 1977, and 1986, respectively. These respective dates correspond to seminar publications by Watson & Crick, Maxam & Gilbert, and Mullis & coworkers. The results shown in the following graph attest to my often stated “power of PCR” as premier method in nucleic acid research, which we’ll see again below in another numerical context.
 Top 5 Cited Papers
During my perusal of the above literature in PubMed generally related to DNA, I thought it would be interesting to find, and share here, which specific papers have the distinction of being most frequently cited. Citations are not available in PubMed, but are compiled in Google Scholar, which led me to these Top 5 that are listed from first to fifth.
Frederick Sanger, the eponymous father of the “Sanger sequencing” method published in 1977, received the 1980 Nobel Prize in chemistry for this contribution. He also received the 1958 Nobel Prize in chemistry for sequencing insulin, and is the only person to win two Nobel Prizes in chemistry. Uber-famous DNA expert Craig Venter is quoted as saying that ‘Fred Sanger was one of the most important scientists of the 20th century,’ [who>
Top 5 Cited Papers
During my perusal of the above literature in PubMed generally related to DNA, I thought it would be interesting to find, and share here, which specific papers have the distinction of being most frequently cited. Citations are not available in PubMed, but are compiled in Google Scholar, which led me to these Top 5 that are listed from first to fifth.
Frederick Sanger, the eponymous father of the “Sanger sequencing” method published in 1977, received the 1980 Nobel Prize in chemistry for this contribution. He also received the 1958 Nobel Prize in chemistry for sequencing insulin, and is the only person to win two Nobel Prizes in chemistry. Uber-famous DNA expert Craig Venter is quoted as saying that ‘Fred Sanger was one of the most important scientists of the 20th century,’ [who> Kenneth J. Livak, PhD
Kenneth J. Livak, PhD
Prof. Bert Vogelstein, MD
Kary B. Mullis, PHD


 Prof. Bert Vogelstein, MD
Prof. Bert Vogelstein, MD Kary B. Mullis, PHD
Kary B. Mullis, PHD



