- CRISPR-Associated Transposase (CAST) from Cyanobacterium Provides the Key for Insertion
- Up to 10 Kilobases of DNA Was Inserted Efficiently into the Target Plasmid by CAST
- Inserting Large DNA Segments with CAST Holds Great Promise for Genetic Engineering
Introduction
Even though there have been multiple blogs posted in the Zone on gene editing using the CRISPR system, interest in this field continues to grow. A PubMed search for the annual number of articles published that have “CRISPR” anywhere in them, as given here for 2013-2018, provides a good metric for assessing this rising interest. The ~4,000 publications published in 2018 translate to an average of ~11 papers per day, or nearly one paper every 2 hours, which is quite impressive.
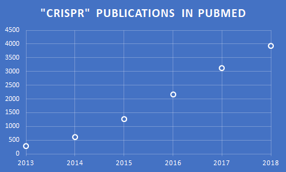 PubMed search and chart by Jerry Zon
PubMed search and chart by Jerry Zon
Most prokaryotes, such as bacteria, rely on the CRISPR-Cas system for adaptive immunity against viruses and mobile genetic elements. Specific sequences of RNAs produced from CRISPR direct Cas effector proteins to seek and destroy nucleic acids from invaders that have complementary target sites. There are multiple types of CRISPR, and each is defined on the basis of its protein composition. Recently, RNA-guided nucleases from types II and V CRISPR systems, Cas9 and Cas12, have revolutionized genome editing by allowing programmed DNA sequence alterations (e.g. see Knott and Doudna).
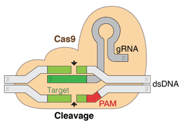 CRISPR-Cas9 system. Taken from commons.wikimedia.org and free to use.
CRISPR-Cas9 system. Taken from commons.wikimedia.org and free to use.
In a typical CRISPR application, Cas9 or Cas12 is directed by its guide RNA to the intended complementary genomic site, flanked by a protospacer-adjacent motif (PAM). This creates a DNA double-strand break (DSB), as depicted here for Cas 9. The outcome of gene editing largely depends on the repair of this DSB by the endogenous pathways in the host cell. Repair by end- joining DNA repair pathways often predominates, and tends to introduce heterogeneous small DNA insertions or deletions (indels) that disrupt a gene’s function. The overall effect is commonly referred to as gene knock-out.
Gene Knock-in Using CRISPR and a Transposase
Despite the proven utility of CRISPR-Cas-based gene knock-out, many applications require site specific gene knock-in of long DNA segments, such as a therapeutic gene or reporter genes. Now, for the first time, long-DNA knock-in has been demonstrated by an MIT team led by Feng Zhang (pictured here), in collaboration with investigators at the NIH. Feng Zhang is well known for his seminal contributions to CRISPR-Cas science and technology, and has been previously featured in the Zone.
 Feng Zhang, a CRISPR “rock star” and cofounder of the CRISPR-related companies Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences.
Feng Zhang, a CRISPR “rock star” and cofounder of the CRISPR-related companies Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences.
This landmark demonstration of gene knock-in led by Zhang, and published as Strecker et al., shows that a CRISPR-associated transposase (CAST) mediates highly efficient, RNA-guided insertion of “cargo” DNA into the bacterial Escherichia coli (E. coli) genome. DNA transposons, also known as “jumping genes,” can move from one genomic location to another, and their relocations and insertions are catalyzed by an enzyme complex called transposase. Incidentally, the study was viewed by more than 23,000 readers of Science during its first two months of availability, according to article usage statistics.
Since 2017, the CAST systems have been discovered through bioinformatics mining of new CRISPR variants, as described by Peters et al., CAST comprises a miniature type I or V CRISPR-Cas encoded within a Tn7-like transposon. The “mini-CRISPR” systems encode protein machineries that either lack the Cas3 nuclease helicase of the type I CRISPR system, or carry inactivating mutations in the catalytic residues of Cas12k of the type V system. Accordingly, they could be competent for target sequence binding, but not for DNA cleavage.
It is speculated that these defective CRISPRs have been “hijacked” by Tn7-like transposons to serve a role other than prokaryotic adaptive defense. Instead, they might increase the evolutionary success of transposons by enabling their RNA-guided spread across bacterial genomes and other mobile DNA elements. The putative RNA-programmable nature of CAST contrasts with the poor target site selectivity of canonical transposases, which insert their cargo DNA semi-randomly.
Strecker et al. established the functionality and underlying mechanism of two CAST loci from cyanobacteria in vivo: Scytonema hofmanni (Sh), pictured here, and Anabaenacylindrica (Ac).
Fluorescent micrograph of the cyanobacteria S. hofmanni; scale bars, 40 mm. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
As depicted here for Sh, each CAST locus is ~20 kilobases, comprising Tn7-like transposase genes (tnsB, tnsC, tniQ), additional cargo genes, and genes encoding a type V-K CRISPR and its effector Cas12k, as well as trans-activating crRNA (tracrRNA), an invariant small RNA cofactor essential for Cas12k.
Schematic of the S. hofmanni CAST locus containing Tn7-like proteins, the CRISPR-Cas effector Cas12k, and a CRISPR array. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
Strecker et al. assayed potential transposition into plasmid DNA in the heterologous host E. coli. As schematically outlined here, a helper plasmid (pHelper) encoding all of the aforementioned CAST protein and RNA components, was delivered into E.coli with a donor plasmid (pDonor) and a target plasmid (pTarget).
Schematic of experiment to test CAST system activity in E. coli. kanR, kanamycin resistance gene; F, forward primer; R, reverse primer. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
Targeted donor integration occurred in one specific orientation, with an efficiency of 13 to 75%. Integration was guided by CRISPR-Cas12k, as it requires tracrRNA, a cognate target, and a 5’ flanking GTN (N=A, G, C or T) protospacer adjacent motif (PAM). Most notably, the insertion sites were clustered unidirectionally downstream the PAM within a narrow window of DNA sequence; 60 to 66 base pairs for ShCAST, and 49 to 56 base pairs for AcCAST, as depicted and discussed below. Donor cargo DNA of up to 10 kilobases was efficiently integrated into the target plasmid.
In Vitro Reconstitution of ShCAST
Strecker et al. state that their data strongly suggests that ShCAST mediates RNA-guided DNA insertion. However, to exclude the requirement for additional cell-host factors, they next sought to reconstitute the reaction in vitro. To do this, they purified all four ShCAST proteins and performed in vitro reactions using pDonor, pTarget, and purified RNA (see Panel A). Addition of all four protein components, crRNA, and tracrRNA resulted in DNA insertions detected by both LE and RE junction PCRs, as did reactions containing the four protein components and sgRNA (See Panel B).
In vitro reconstitution of an RNA-guided transposase. (A) Schematic of in vitro transposition reactions with purified ShCAST proteins and plasmid donor and targets. (B) RNA requirements for in vitro transposition. pInsert was detected by PCR for LE and RE junctions. All reactions contained pDonor and pTarget. Schematics indicate the location of primers and the expected product sizes for all reactions. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
Model for RNA-guided DNA Transposition
Strecker et al. demonstrated that CRISPR-Cas systems associated with Tn7-like transposon mediate RNA guided DNA transposition, and elucidated the underlying molecular mechanism. ShCAST mediates unidirectional insertions in a narrow window downstream of the target, as depicted here. The researchers also state that, although ShCAST and AcCAST exhibit similar PAM preferences, one notable difference is that their respective positions of insertion, relative to the PAM, differ by 10 to 11 bp, which roughly corresponds to one turn of DNA. “Deeper exploration of microbial genomes is expected to uncover CAST systems with a range of diverse properties, including targeting preference and activity across different conditions” according to Strecker et al. They further observe that targeted DNA insertion by ShCAST results in the incorporation of LE and RE elements (see also the schematic of the S. hofmanni CAST locus) and is therefore not a “scarless” integration method.
Model for RNA-guided DNA transposition. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
Future Prospects
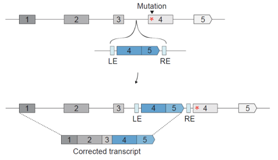
Strecker et al. foresee that one potential generalizable strategy for the use of CAST in a therapeutic context would be to insert corrected exons into the intron before the mutated exon, as depicted here.
Potential strategy for CAST-mediated gene correction. Replacement of a mutation-containing exon by targeted DNA insertion. Taken from Strecker et al. Science, 2019 Jul 5;365(6448):48-53. Copyright © 2019 The Authors, with permission.
They add that CAST could also be used to insert transgenes into “safe harbor” loci, which Sadelain et al. propose as “chromosomal locations where therapeutic transgenes can integrate and function in a predictable manner without perturbing endogenous gene activity and promoting cancer.” Finally, CAST is envisaged by Strecker et al. as being used downstream of endogenous promoters so that the expression of transgenes of interest can benefit from endogenous gene regulation.
However, some caveats were noted by Strecker et al. For example, their analysis indicated that ShCAST is specific but, under conditions of overexpression, can integrate at non-targeted sites in the E. coli genome via Cas12k-independent mechanisms, and this guide-independent integration seems to favor highly expressed genes. They also observed non-targeted insertions into pHelper in E. coli that were independent of Cas12k. Strecker et al. believe that “[f>
uture protein engineering of the transposase components could improve the targeting specificity of CAST systems.”

his work identifies a function for CRISPR Cas systems beyond adaptive immunity that does not require Cas nuclease activity and provides a strategy for targeted insertion of DNA without engaging homologous recombination pathways, with a particularly exciting potential for genome editing in eukaryotic cells.”
I agree with this future-view of CAST, but feel compelled to note that all previously pursued nucleic acid-based therapeutic strategies, such as antisense, siRNA, and CRISPR base-editing, have encountered problems with specificity/off-target effects. These issues, as well as effective delivery of all nucleic acid-based agents, continue to present a big challenge.
As usual, your comments are welcomed.