- Prof. Eric Kool Continues to Innovate Chemistries to Manipulate RNA
- His Versatile Chemical Toolbox Opens the Door for New Applications
- Reversible Cloaking of RNA for Temporal Control of CRISPR Gene-Editing Is His Latest Achievement
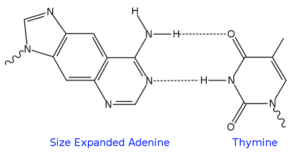
Eric Kool, a professor of chemistry at Stanford University, is very well known in the field of nucleic acids research. According to citation statistics in Google Scholar, Kool’s most impactful publications to date have dealt with the elucidation of hydrogen bonding and base stacking in duplex DNA. This insightful research allowed him to design a new molecular class of a genetic-pairing system in which a native DNA backbone is retained, but all four base pairs are replaced by new, larger pairs. The base pairs include size-expanded analogs of A, G, C and T wherein a benzo moiety has been fused, as pictured here for size-expanded A.
 Taken from commons.wikimedia.org and free to use.
Taken from commons.wikimedia.org and free to use.
 By following and learning from Kool’s publications for quite a while, it is apparent to me that his interests have recently expanded (pun intended) from DNA to RNA. This expansion from deoxy- to ribonucleic acids has led to a number of innovative chemical methods for modifying RNA in order to manipulate RNA function in controlled ways. As will be apparent from this blog, which covers past to present publications, Kool’s enlargement of the “chemical toolbox” for modifying RNA opens the door for new applications, including temporal control of CRISPR-based gene-editing.
By following and learning from Kool’s publications for quite a while, it is apparent to me that his interests have recently expanded (pun intended) from DNA to RNA. This expansion from deoxy- to ribonucleic acids has led to a number of innovative chemical methods for modifying RNA in order to manipulate RNA function in controlled ways. As will be apparent from this blog, which covers past to present publications, Kool’s enlargement of the “chemical toolbox” for modifying RNA opens the door for new applications, including temporal control of CRISPR-based gene-editing.
RNA Cloaking by Reversible Acylation
 NAI-N3 PubChem Sketcher structure by Jerry Zon.
NAI-N3 PubChem Sketcher structure by Jerry Zon.
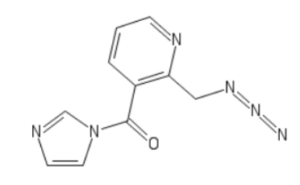
In February 2018, Kool’s lab reported a novel and versatile strategy for covalent acylation and de-acylation of RNA. The approach is distinct from a prior siRNA photo-caging method in two important ways: it is carried out post-synthetically in a single step, and it is reversed chemically rather than by light. in contrast to previous RNA acylation reactions, Kool’s group showed that the reaction of RNA with an azide-bearing acylating reagent, abbreviated NAI-N3 and shown here, can be carried out in high yields with multiple acyl blocking groups appended to each strand.
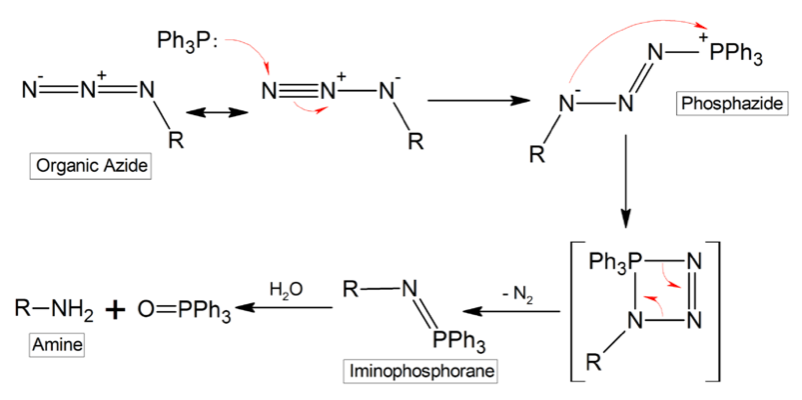
The researchers further showed that this polyacylation (“cloaking”) can block various RNA functions such as folding, small molecule binding, hybridization, and enzyme recognition. At a subsequent time, the unmodified RNA with native activity can be restored by bioorthogonal deacylation with a water-soluble phosphine. The phosphine reactant triggers a Staudinger reduction of the azide groups, as depicted here for Ph3P, thus resulting in spontaneous loss of acyl groups (“uncloaking”). Consequently, this new cloaking and uncloaking strategy enables chemical control of RNA.
The methodology is exceptionally simple and mild, and functions both with chemically synthesized RNAs and longer transcribed ones. Readers interested in the details of this cloaking and uncloaking method and examples of its application can consult the original publication and supplementary material at this link.
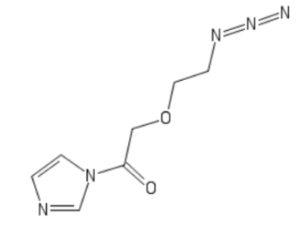
Kool’s lab has extended this methodology in two directions. The first involves cloaking with an acyl moiety bearing a photo-labile group to enable light-induced uncloaking of RNA, as described in a previous blog in the Zone, while the second involves cloaking with relatively simple azido-functionalized alkanoyl acylating agents, such as the imidazoyl (Im) compound shown here.
The polyacylation (cloaking) procedure simply involved the reaction of 6 uM RNA with 0.8 M acylating probe for 10 h at 10 °C in a total volume of 100 uL. The resultant cloaked RNA was then purified via ethanol precipitation. The deacylation (uncloaking) conditions were likewise very simple, and employed incubation of 1 uM RNA in 300 mM Tris-HCl buffer at pH 7.5 containing 20% DMSO with 20 mM water-soluble 2-(diphenylphosphino) ethylamine for 4 h at 37 °C in a total volume of 100 uL. The resultant RNA was purified via an ethanol precipitation procedure identical to the one utilized after RNA cloaking. Mechanistically, uncloaking is the result of Staudinger reduction of N3 to NH2, which then attacks the carbonyl, leading to displacement of 2’-OH.
Hydrophobic Functionalization of RNA
Some major advantages of the aforementioned polyacylation strategy are that it can be applied in a single step to unmodified RNA of any length, and it does not require solid-phase nucleic acid synthesis. In October 2018, Kool’s lab reported a new variation of this method to enable functionalization of RNA with hydrophobic groups that are either necessary for certain applications, such as dye tagging, or might enable entirely new applications of modified RNA.
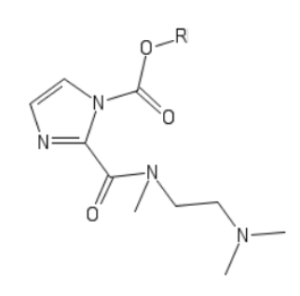
To overcome this limitation, they developed a highly water-soluble imidazole leaving group that would enable functionalization of RNA with hydrophobic moieties. As depicted here for the reagent abbreviated MIC, a tertiary amine was chosen to be appended to the imidazoyl carbonyl via an N-methyl amide bond.
The ability of this generalized structure to provide a water-soluble imidazole leaving group and enable functionalization of RNA with nonpolar acyl groups was assessed using a range of hydrophobic groups, which were introduced by reactions of the corresponding chloroformates, RO(CO)Cl. The investigated compounds were chosen to represent alkyl groups (R) of varied length and branching, as well as a benzyl group (PhCH2). To functionalize RNA, the compounds were dissolved in water at a concentration of 0.1 M and incubated with a 15-mer RNA oligonucleotide (CAUAGGUCUUAACUU) at 10 μM for 16 h.
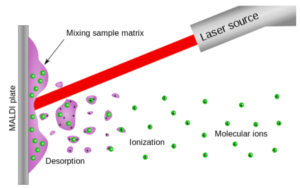
Functionalized RNA was purified by precipitation or filtration with a molecular weight cutoff-filter, with yields varying widely depending on substitution. The relative degree of RNA functionalization was analyzed by MALDI-TOF mass spectrometry, depicted here in a generalized scheme.
In time-of-flight (TOF) mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy (red) absorbing matrix (pink) to create ions (green) from large molecules with minimal fragmentation. Taken from commons.wikimedia.org and free to use.
Overall, the reaction efficiency was found to be strongly influenced by the substitution pattern around the α- carbon. Reaction of the 15-mer RNA with the methyl derivative resulted in a median of 2−3 added groups per strand, whereas increasing the substitution decreased the number of groups to 1−2 for the ethyl derivative, 0−1 for the isopropyl compound, and virtually no functionalization with the tert-butyl (t-Bu) derivative. The neopentyl compound contains an additional methylene between the t-Bu group and carbamate, and resulted in increased levels of reaction compared to the t-Bu compound.
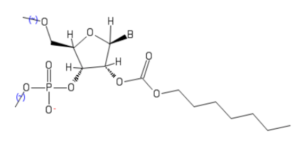
Interestingly, increasing the alkyl chain length seemed to dramatically increase functionalization, as seen for the n-heptyl derivative (depicted here), which exhibited a median of 4 added groups per RNA. The Kool group hypothesized that hydrophobic groups such as n-heptyl might give the RNA extraordinary properties, such as improved solubility in organic solvents. Control unmodified RNA was mixed with water, ethanol, or acetonitrile at a theoretical concentration of 4 μM, and solubility was measured by RNA absorbance at 260 nm. Only when mixed with water was unmodified RNA absorbance observed.

Polyacetate and Polycarbonate RNA
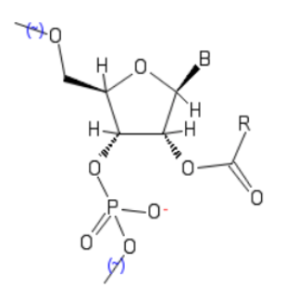
In further exploring the scope and utility of polyacetylation and cognate modification of RNA, in July 2019, Kool’s lab reported the acylation of 2’-OH groups with “the smallest 2-carbon acyl groups on RNA”—namely, 2’-O-acetyl and 2’-carbonate groups, as depicted here. Chief goals were to obtain increased levels of modification using readily available reagents, and retain inhibition of RNA function.
To measure reactivity for esterification of RNA, the researchers reacted acetylimidazole (AcIm) with RNA at varying concentrations and times. It was found that fresh preparation of AcIm in dry DMSO was superior to the use of older bottles of AcIm, as these are significantly hydrolyzed. The half-life of AcIm in 25 mM aqueous phosphate buffer (pH 7.4) was determined to be ~30 min at room temperature. After reaction, RNAs were analyzed by MALDI-TOF mass spectrometry. The results showed that AcIm does react with RNA in high yields, yielding up to 89% acetylation of nucleotides in a concentration-dependent manner. For a representative 18-mer RNA, the median level of acylation was found to be distributed among 10−14 acetyl groups.
Kool’s group similarly tested acylation of RNA by the methyl imidazole carbamate compound (MIC) mentioned above, which has a half-life of 24 h in 25 mM phosphate buffer (pH 7.4) at room temperature. The results in this case also showed high levels of reactivity with the 18-mer RNA, again yielding adducts up to 89% of nucleotides depending on concentration, but requiring longer incubation times compared to AcIm. The median level of acylation was found to be distributed among 4−6 carbonate groups.
To test the RNA selectivity of these acylation reactions, Kool’s group compared reactions of AcIm with DNA and RNA. The results showed only minor reaction with DNA, confirming the need for 2′-OH groups as nucleophiles in the reactions, and ruling out major reaction with exocyclic amine groups. They also tested reactions of RNAs in duplex with DNA, and acylation yields were reduced markedly, suggesting that the 2′-OH groups in double-stranded helical RNA are not readily accessible even to these small reagents. Kool’s group applied this differential reactivity to map RNA using a method called SHAPE.
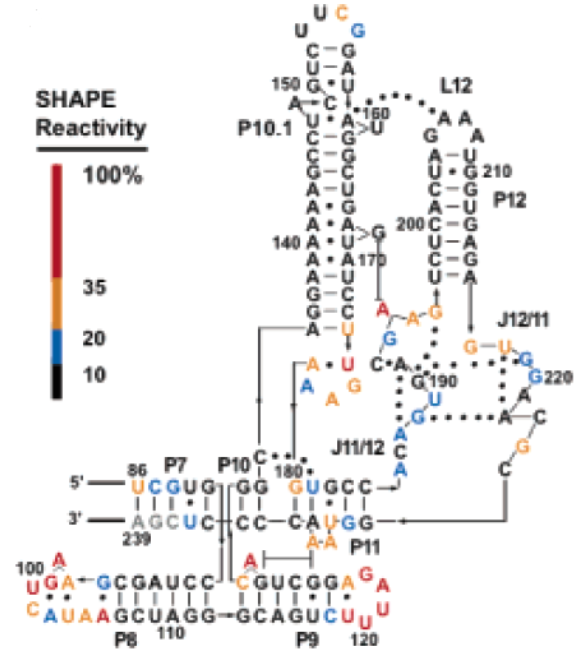
Selective 2′-hydroxyl acylation analyzed by primer extension, or SHAPE, exploits reagents that preferentially modify the backbone of RNA in structurally flexible regions. Adduct formation is quantified for each nucleotide in a given RNA by extension of a complementary DNA primer with reverse transcriptase, and comparison of the resulting fragments with those from an unmodified control. SHAPE therefore reports on RNA structure at the individual nucleotide level. This data can be used as input to generate highly accurate secondary structure models, as shown here for base pairing and tertiary interactions for the specificity domain of Bacillus subtilis RNase P.
Reversible RNA Acylation for Control of CRISPR-Cas9 Gene Editing
Given the extent and intensity of interest in CRISPR-related applications, which have been discussed in previous blogs in the Zone, investigation of reversible RNA acylation for control of CRISPR-Cas9 gene editing is especially exciting. Initial results were described in the poster shown below, which was presented at the 2018 International Roundtable (IRT) on Nucleosides, Nucleotides and Nucleic Acids held at UC San Diego. The presenter, Maryam Habibian, recently informed me that an updated version of these studies has been accepted for publication in J. Amer. Chem. Soc.
In brief, the poster indicates that the NAI-N3 acylating reagent mentioned above efficiently acylates CRISPR single guide RNAs (sgRNAs) in 20 min in buffer. Cloaked, poly-acylated sgRNA was completely inactive in the DNA-cleaving activity of Cas9 in vitro, and was quantitatively restored after treatment with a phosphine. Delivery of cloaked sgRNA and Cas9 mRNA (available from TriLink as a catalog item) into HeLa cells was enabled by the use of CART cationic polymers (see previous Zone blog), which outperformed commercial transfection reagents. Genomic DNA cleavage in the cells by CRISPR-Cas9 was efficiently restored after treatment with the phosphine to remove the blocking acyl groups from the sgRNA. These results were said to highlight the utility of reversible RNA acylation as a novel method for temporal (i.e. time-based) control of genome-editing function.
Your comments are welcomed, as usual.