- Khorana’s Dream of Synthesizing a Gene from Hand-Made Oligos
- Caruther’s Dream of Automating Oligo Synthesis
- Venter’s Dream of Fully Automating Gene Synthesis
- Who’s Dreaming About What’s Next?
 This blog acknowledging National DNA Day on April 25th deals with dreams of various sorts, but mainly with gene synthesis, which was only a dream in the 1950s and is now achievable in a way few dreamed possible even a few years ago.
This blog acknowledging National DNA Day on April 25th deals with dreams of various sorts, but mainly with gene synthesis, which was only a dream in the 1950s and is now achievable in a way few dreamed possible even a few years ago.
Before I get to DNA gene-dreams that did come true, I want to briefly mention two other dream-like anniversaries. First is the fact that my blog is now beginning its 4th year—yeh!—after its inaugural posting in April 2013 to celebrate 60 years since Watson & Crick’s famous publication of DNA’s helix structure as the fundamental basis for genetic material. Second is this year being TriLink’s 20th anniversary—yeh!—as a leading provider of modified nucleic acids, which co-founders Rick Hogrefe and Terry Beck likely view as their business dream come true. But I digress…
The First Dreamer and Doer
 Har Gobind Khorana. Taken from newsoffice.mit.edu.
Har Gobind Khorana. Taken from newsoffice.mit.edu.
I don’t know when or who actually first dreamed of synthesizing a gene, but it could very well have been H. Gobind Khorana, who shared the Nobel Prize in Physiology or Medicine in 1968 with Robert W. Holley and Marshall W. Nirenberg "for their interpretation of the genetic code and its function in protein synthesis.” More to the point of his DNA dream, Khorana and coworkers at the University of Wisconsin, Madison published the first-ever synthesis of a gene, namely that encoding alanine tRNA from yeast, in Nature in 1970.
By exploiting the natural ability of polynucleotides to align by base pairing and using polynucleotide kinase and ligase, chemically synthesized segments were combined into the double-stranded DNA corresponding to this tRNA.
Having read a number of the many early DNA synthesis papers by Khorana, I felt an almost tangible sense of the huge amount of lab work by a small army, as it were, of highly skilled chemists necessary for achieving this historic synthesis of a relatively small gene. However, that was destined to change dramatically, largely due to Marvin H. Caruthers, who was among the twelve “coworkers” referred to above, and was then a youngish 30-year-old postdoc with his own dreams about DNA.
“Gene Machine” Dreams: Taking the Manual & the Art Out of DNA Synthesis
Manual synthesis of DNA “back in the day” was not only slow, tedious work, but also somewhat of an art best performed by chemists with so-called “great hands” in the lab. Consequently, it was inevitable that various approaches were pursued to automate synthesis of oligos, from which genes were constructed and—more importantly—allowed non-chemists to easily produce these oligos.
For historical purposes, a number of companies launched what were popularly referred to as “gene machines,” a misleading distinction since the output was not genes per se but oligo components thereof. Variously designed gene machines that I found in 1980s vintage publications included Analysteknik NUCSYN (1982), Vega Biochemicals DNA Synthesizer (1982), ABI Model 380A (1983), Bachem Gene Machine (1985), and Biosearch SAM I (1985). Others can be read about in a 1990 article from The Scientist.
By far the most successful of these early gene machines was that based on Caruther’s phosphoramidite chemistry commercialized by ABI. This machine was presciently discussed by Leroy Hood and Mike Hunkapiller at Caltech in a forward-looking 1983 perspective entitled Biotechnology and Medicine of the Future, which was followed by their related 1984 article in Nature coauthored with Caruthers and others.
My two-sentence personal story related to this era was that in 1982 I flew to Vega Biochemicals and then on to ABI to assess each company’s oligo synthesizer for purchase. I opted for the latter in large part because of its potential to automatically synthesize various modified oligos—notably phosphorothioates—as published in JACS in 1984—with my good—and now famous—friends Wojciech J. Stec and William Egan. This drew me into what would become my dream—the new field of oligonucleotide therapeutics.
Bigger Dreams on a Smaller Scale
The 1990s ushered in the advent of new technologies for photolithographic and ink-jet methods for parallel synthesis of truly huge numbers of oligos on very small scales for high-density genotyping and sequencing-by-hybridization, as reviewed elsewhere. This paradigm-shift also led to investigations of how oligos cleaved from such arrays (aka “chips”) could be used for gene synthesis.
One notable achievement was published in 2007 by Jacobson and coworkers at MIT, who reported the fabrication of a multi-chamber microfluidic device and its use in carrying out the syntheses of several DNA constructs. Using conventionally prepared oligos, genes up to 1-kb in length were synthesized (as depicted below) in parallel at minute starting oligonucleotide concentrations (10–25 nM) in four 500-nL reactors, which were 10- to 100-times lower than those utilized in conventional gene synthesis.
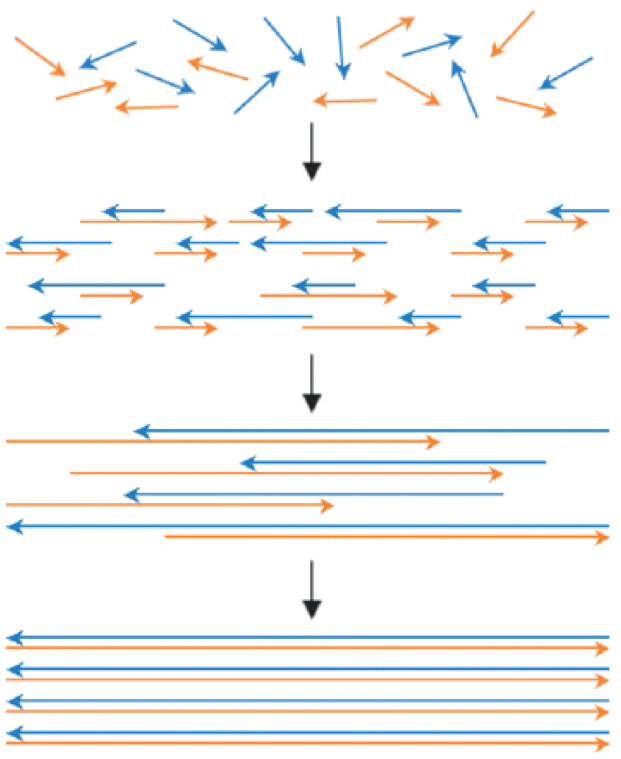 Gene synthesis by polymerase construction and amplification (PCA) wherein multiple rounds of oligo annealing and extension generate successively longer DNA assemblies from a starting pool of construction oligos, typically
Gene synthesis by polymerase construction and amplification (PCA) wherein multiple rounds of oligo annealing and extension generate successively longer DNA assemblies from a starting pool of construction oligos, typically
Fully Automated Gene Synthesis: "Push Button" Genes
In my opinion, defining hallmarks of Craig Venter’s scientific dreams, so to speak, are that they are unconventional and very big—e.g. (1) shotgun sequencing human genomes by Celera Corporation faster and cheaper than the publically funded effort, (2) first-ever complete chemical synthesis of a genome, and (3) increasing human longevity—albeit corporate work-in-progress.
Consequently, I was very pleased, but not too surprised, to learn a while back that one of Venter’s next dreams was to form (within Synthetic Genomics Inc.) a subsidiary named SGI-DNA to commercialize an instrument for fully automated gene synthesis. This formidable and daunting goal has been recently realized by launch of the BioXp™ 3200 System that is loosely referred to as the world’s first “DNA Printer.”
Details can be read elsewhere, but a key scientific element worth noting here is that the methodology for assembling otherwise conventionally synthesized oligos is based upon the so-called Gibson Assembly strategy originally developed for (2) referenced above and depicted below. The BioXp™ 3200 System allows users to simply input desired gene sequences into an SGI-DNA website and then receive a plate of corresponding synthetic oligonucleotides for use with other required reagents. In conjunction with the newly introduced cloning module, the operator pushes the "go button”, if you will, and voilà, a plasmid-encoded gene is provided with high efficiency!

When I inquired about error rates, Claudia H. Alvarez of SGI-DNA replied that, “specific error rate per construct and overall sequence fidelity of a cloned product is dependent upon various factors including but not limited to: oligo pool quality, error correction and construct size. Currently, SGI-DNA provides [its> customers with a table that depicts the number of clones to pick based on construct size.”
Number of Full Length Clones Recommended to Sequence to Identify an Error Free Clone
| DNA fragment size | # Clones to pick (full length) |
| < 1000 bp | 2 to 4 |
| 1000 – 1500 bp | 4 to 8 |
| 1501 – 1800 bp | 8 to 10 |
Don't Stop Dreaming
I chose this concluding section subtitle partly because it’s the name of a song by a group called Celebration—no joke—that encapsulates my parting thought on the above dreams of DNA, which is that we shouldn’t stop dreaming of what could be.
I’m confident that some of what may now be viewed as either wishful thinking or just a dream will be realized, just as Khorana’s once seemingly impossible dream of synthesizing a gene from hand-made oligos was not only realized, but is now as simple as pushing a button on the BioXp™ 3200 System.
Some intrepid DNA dreamers worth reading about are Twist Bioscience, which is “Rewriting DNA Synthesis with the Power of Scale”, and Molecular Assemblies, which is “Enabling a New Era in Biotechnology” by developing a cost-effective, sustainable approach for enzymatic synthesis of long Eco-DNA to replace chemicals.
I welcome you to share, by commenting here, other DNA-dream efforts you may know about, or share your own dreams about DNA.






