- Pancreatic Cancer is the Third Leading Cause of Cancer Deaths in the US
- Early Diagnosis is Needed in Order to Increase Survival Rates
- Analysis of Cell-Free DNA Biomarkers in Blood May Fulfill This Need
Introduction
The pancreas is an organ located behind the stomach and in front of the spine. It is shaped a bit like a fish with a wide head, a tapering body, and a narrow, pointed tail. In adults it's about 6 inches long, but less than 2 inches wide. The head of the pancreas is where the stomach meets the duodenum (the first part of the small intestine). The body of the pancreas is behind the stomach, and the tail is next to the spleen. Exocrine cells make enzymes that are released into the intestines to help digest foods, and endocrine cells secrete hormones that help control blood sugar levels.
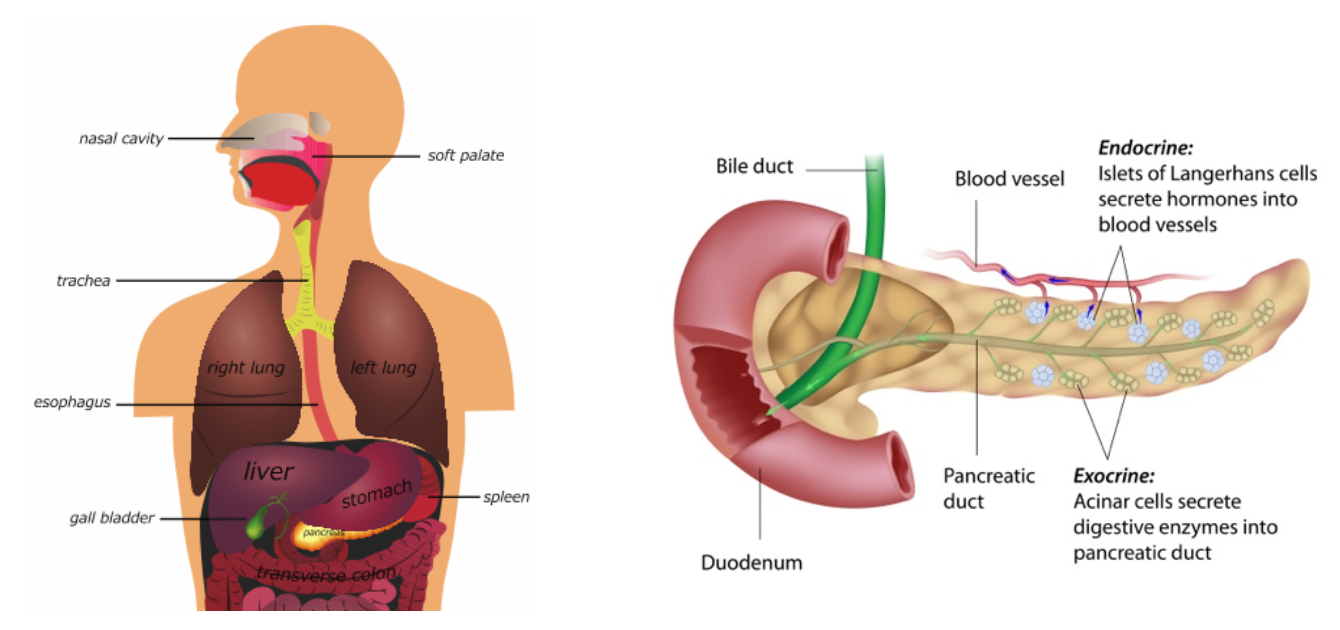 Illustration of the pancreas and surrounding organs. // Illustration of exocrine and endocrine pancreas.
Illustration of the pancreas and surrounding organs. // Illustration of exocrine and endocrine pancreas.
The American Cancer Society estimates that in 2019 in the United States, about 57,000 people (30,000 men and 27,000 women) will be diagnosed with pancreatic cancer (PC), and about 46,000 people (24,000 men and 22,000 women) will die from it. These statistics not only make PC the nation’s third leading cause of cancer deaths after cancers of the lung and colon, but also put it on track to overtake colon cancer within a decade. Three-fourths of people who develop PC die within one year of diagnosis, and only about one in 10 live five years or longer. The main reason for these grim survival statistics is that symptoms for PC may not be apparent in the early stages, and patients may therefore be misdiagnosed. Unfortunately, while there are screening tools such as mammographies for breast cancer or colonoscopies for colon cancer, there are no comparable diagnostics for the screening of PC in seemingly healthy individuals, when the cancer would be most amenable to cure.
However, as will be evident from the information provided in this blog, significant progress is being made toward early detection using cell-free DNA (cfDNA) biomarkers. Such biomarkers are especially attractive because they could allow for early detection in readily available body fluids. These “liquid biopsies” would be much easier and safer than current procedures utilized for pancreas biopsy, which are extremely invasive due to the sequestered location of this organ. One of these procedures, brush biopsy during endoscopic retrograde cholangiopancreatography (ERCP), is depicted here.
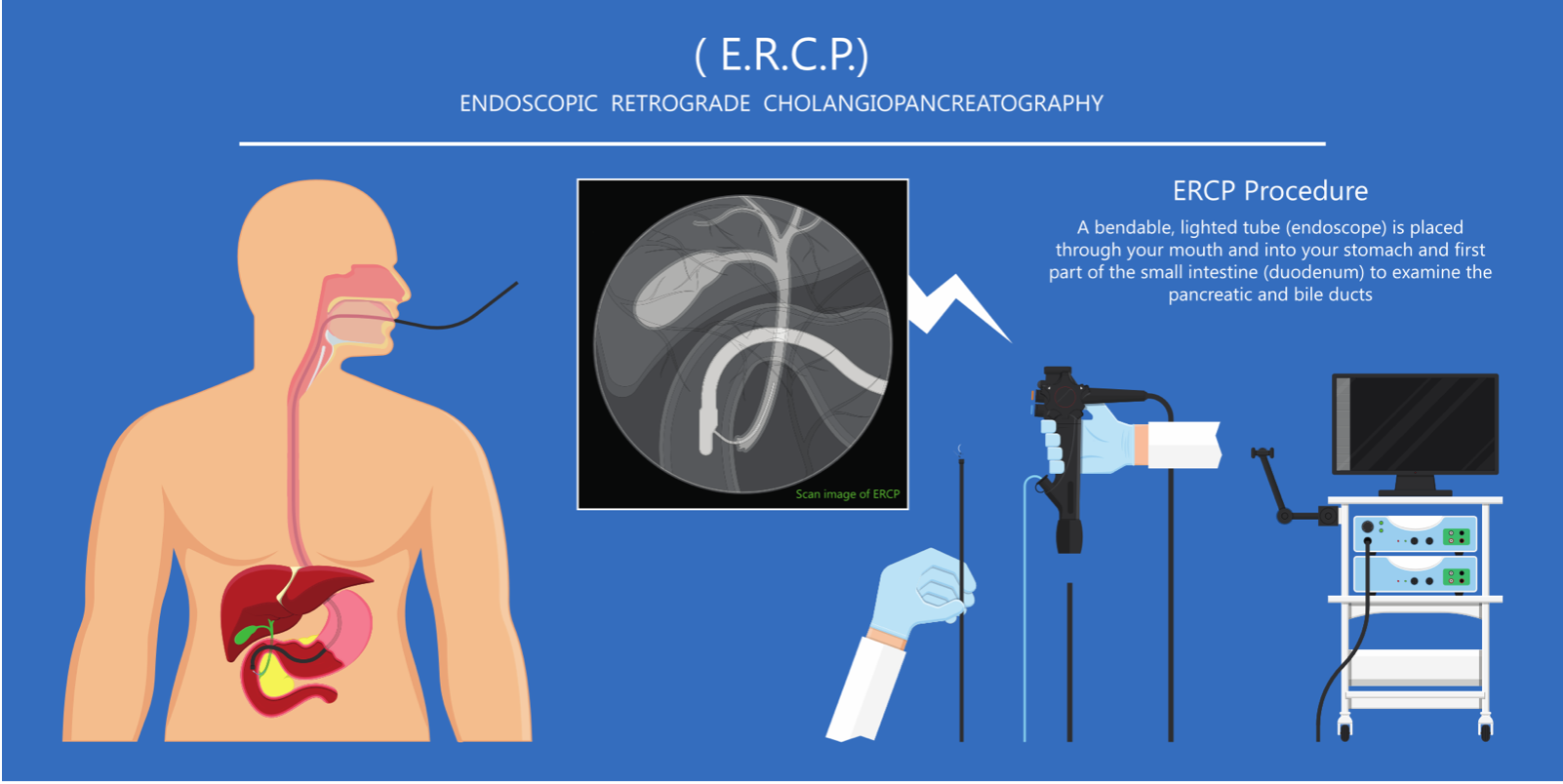
Brush biopsy: a long, thin, lighted tube called an endoscope is inserted through the mouth into the first part of the small intestine. The doctor then places a tiny brush through the endoscope and into the pancreatic or bile ducts. The brush rubs off some cells for removal and lab tests.
Publications About PC and Biomarkers
The potential advantages and need for liquid biopsies in early detection of PC have stimulated a large amount of research aimed towards achieving that goal. To assess the extent of this interest, I searched the freely available National Library of Medicine/National Institutes of Health PubMed database for publications in this area. Searching for publications during 2008-2018 that have titles and/or abstracts containing the phrase “pancreatic cancer” and the word “biomarkers” led to about 1,000 articles. The chart of these items grouped by year of publication, as depicted here, clearly showed an upward trend, with nearly 200 articles in 2018 alone.
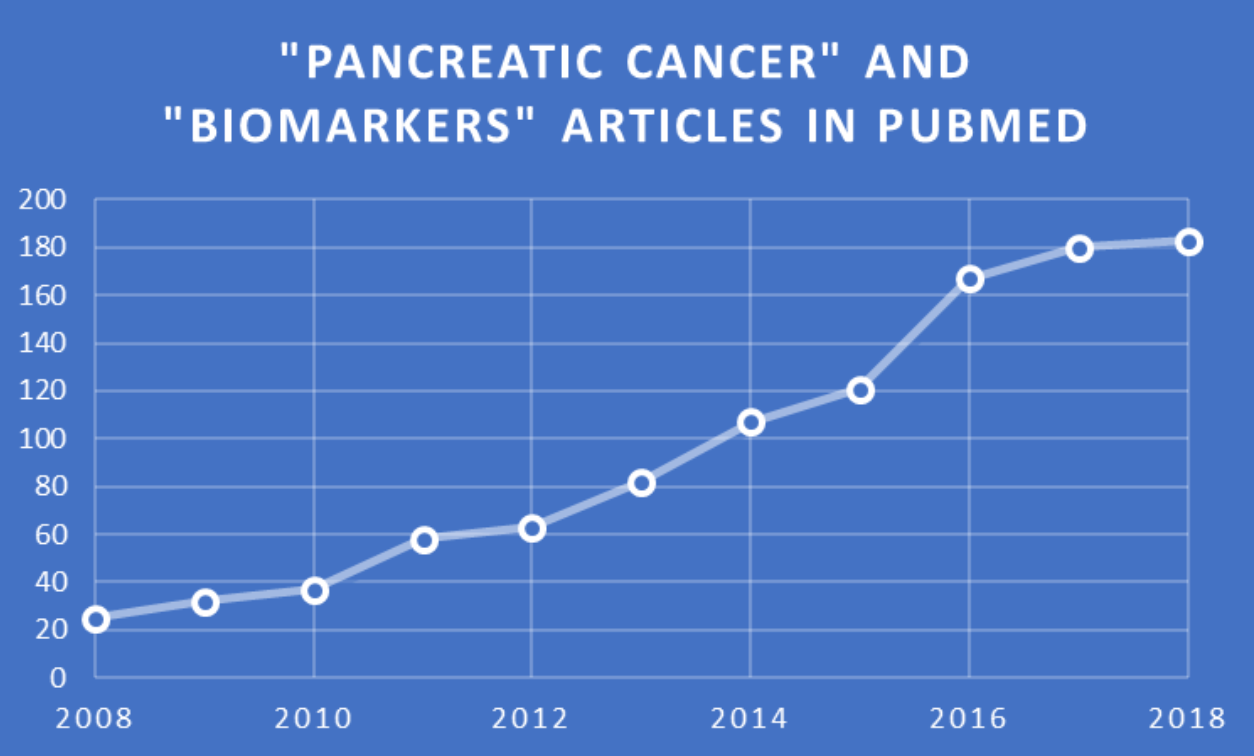 PubMed search results and chart by Jerry Zon.
PubMed search results and chart by Jerry Zon.
Biomarkers are generally comprised of either nucleic acids, proteins, metabolites, or combinations thereof. In keeping with the Zone’s focus on what is trending in nucleic acids research, the aforementioned search was repeated, but “biomarkers” was replaced by the phrase “cell-free DNA,” which led to 43 articles (including reviews) published from 2012 to present. All of these PC/cfDNA publications can be accessed at this link, as the remainder of this blog will offer short synopses of five papers that, in my opinion, collectively indicate significant progress toward early detection of PC using cfDNA biomarkers.
Deciphering DNA Methylation Signatures of PC

In a September 2019 publication, Natale et al. point out that, while liquid biopsies are convenient and minimally invasive, their utilization for the development of cfDNA methylation biomarkers for PC is challenging. In particular, the very low amounts of PC-specific cfDNA remain a limiting factor. Achieving robust data analysis therefore requires for investigations to be restricted to simultaneous analysis of only a few gene targets from a current list of about 20 methylated cfDNA genes for PC, as single epigenetic markers are seldom sufficient to distinguish PC from benign or pre-neoplastic conditions.

Natale et al. conclude that the decreasing cost of next-generation sequencing (NGS) technologies offers an appealing option for prediction models, and “early diagnosis has the potential to allow PC prognosis prediction, tumor-stage monitoring, and provide personalized therapeutic strategies for patients suffering from PC”. Other researchers have indeed leveraged NGS for this purpose, as outlined in the next section.
Enrichment of Short Mutant cfDNA for Enhanced Detection of PC
In a March 2019 publication, Liu et al. addressed the utilization of limited amounts of cfDNA for PC detection by NGS. First, a single-strand cfDNA library preparation and hybrid-capture-based method for sequencing (SLHC-seq) was developed for the analysis of degraded cfDNA fragments. The degraded nature of cfDNA results from exposure to extracellular enzymes. In addition, cfDNA is highly diverse in terms of its cellular origin, meaning that conventional PCR is not sensitive enough to capture the full diversity of these molecules, particularly with regards to degraded fragments with nicks in either strand.



Schematic representation of the nucleosome, the smallest subunit of the chromatin. The DNA (grey) is wrapped around the nucleosomal core proteins, the histones H2A (yellow), H2B (red), H3 (blue), and H4 (green). The nucleosomal linker is the section of the DNA that is not wrapped around the core. It is normally attached to the histone H1 (not shown). The total length of the DNA per nucleosome is ~180 bp. Image from commons.wikimedia.org.
KRAS is a gene that acts as an on/off switch in cell signaling. When KRAS functions normally, it controls cell proliferation; when it is mutated, negative signaling is disrupted. Thus, cells can continuously proliferate, and often develop into cancer. Liu et al. note that the detection of KRAS mutations in the plasma of PC patients has been studied for a long time, but with significant bias towards late-stage patients. Using SLHC-seq, KRAS hotspot mutations were detected in over 70% of patients in this cohort, which comprised 66% of patients with precancerous or early-stage disease. No hotspot mutations were detected in the circulation of healthy individuals.
Liu et al. concluded that “the performance of SLHC-seq in identifying ctDNA was highly comparable to that of ddPCR-based methods or NGS with ultra-deep sequencing depth.” Importantly, “the high sensitivity and reliability of SLHC-seq make it highly suitable for early cancer detection and intervention, and its ability for large-scale parallel sequencing of multiple and broad genomic regions also enables the quantitative surveillance of tumor-derived mutations that may evolve during early tumorigenesis, tumor progression, and drug resistance.”
Mutant KRAS in Exosome-Derived DNA (exoDNA) vs. cfDNA in Early-Stage PC

Allenson et al. therefore tested the potential of exoDNA as an additional blood-based compartment which may be complementary to cfDNA in the diagnosis and therapeutic stratification of patients with PC. More specifically, they compared exoDNA to cfDNA in liquid biopsies of patients with pancreatic ductal adenocarcinoma (PDAC), which constitutes 90% of all PC.
Exosomes were isolated using serial ultracentrifugation and characterized with electron microscopy, flow cytometry, and particle analysis. CfDNA was isolated using a commercially available kit. Droplet digital polymerase chain reaction (ddPCR), depicted here in a simplified generic form, was applied to detect KRAS mutations with a commercially available multiplex assay.

A cumulative series of 263 individuals were studied, including a discovery cohort of 142 individuals: 68 PDAC patients of all stages; 20 PDAC patients initially staged with localized disease, with blood drawn after resection for curative intent; and 54 age-matched healthy controls. A validation cohort of 121 individuals (39 cancer patients and 82 healthy controls) was studied to validate KRAS detection rates in early-stage PDAC patients. Primary outcome was circulating KRAS status as detected by droplet digital PCR. Secondary outcomes were disease-free and overall survival.
KRAS mutations in exoDNA were identified in 7.4%, 66.7%, 80%, and 85% of age-matched controls, localized, locally advanced, and metastatic PDAC patients, respectively. By comparison, mutant KRAS cfDNA was detected in 14.8%, 45.5%, 30.8%, and 57.9% of these individuals, respectively. Higher exoKRAS mutant allele frequencies (MAFs) were associated with decreased disease-free survival in patients with localized disease. In the validation cohort, mutant KRAS exoDNA was detected in 43.6% of early-stage PDAC patients and in 20% of healthy controls.
Allenson et al. concluded that exosomes are a distinct source of tumor DNA that may be complementary to other liquid biopsy DNA sources. A higher percentage of patients with localized PDAC exhibited detectable KRAS mutations in exoDNA than previously reported for cfDNA. A substantial minority of healthy samples demonstrated mutant KRAS in circulation, dictating careful consideration and application of liquid biopsy findings, which may limit its utility as a broad cancer-screening method.
Concluding Comments
As I am definitely not an expert in cancer diagnostics, I thought it would be appropriate to conclude with comments from bone fide experts. Among the aforementioned 43 articles on PC, I noticed a publication titled “Liquid biopsies in pancreatic cancer” by Kamyabi et al. in Expert Reviews in Anticancer Therapy. The summary expert opinion offered is the following:
“In pancreatic cancer where tissue samples are limited and repeated tissue biopsies are mostly invasive and infeasible, liquid biopsies opened a new window for tumor diagnosis, molecular stratification, and treatment monitoring.”
After researching the literature to write this blog, I am of the opinion that the “new window” referred to by Kamyabi et al. looks out onto liquid biopsies involving cfDNA analysis for improved diagnosis of PC, including early detection, which is indeed a bright prospect.
Your comments are welcomed, as usual.
Addendum
After writing this blog, Science Daily reported that a new blood test in development has shown ability to screen for more than 20 types of cancer, including PC, with a high degree of accuracy. Data for this test, which uses NGS to detect methylation patterns in cfDNA, were presented during a recent session at the European Society for Medical Oncology (ESMO) 2019 Congress.
In the study, investigators analyzed cfDNA in 3,583 blood samples, including 1,530 from patients diagnosed with cancer and 2,053 from people without cancer. The overall specificity was 99.4%, meaning only 0.6% of the results incorrectly indicated that cancer was present. The sensitivity of the assay for detecting pre-specified high-mortality cancers (the percent of blood samples from these patients that tested positive for cancer) was 76%. For 97% of samples that returned a tissue of origin result, the test correctly identified the organ or tissue of origin in 89% of cases.
Additional “late breaking news” was reported in Nature magazine by Aykut et al. on a causal link between a fungal mycobiome and promotion of pancreatic oncogenesis. These researches showed that fungi migrate from the gut to the pancreas, and that this is implicated in the pathogenesis of PDAC. These tumors in humans and mouse models displayed an increase in fungi of about 3,000-fold compared to normal pancreatic tissue. The composition of the mycobiome of PDAC tumors was distinct from that of the gut or normal pancreas. Specifically, the fungal community that infiltrated PDA tumors was markedly enriched for Malassezia (depicted here) in both mice and humans.
Aykut et al. also discovered that ligation of mannose-binding lectin (MBL), which binds to the glycans of the fungal wall to activate the complement cascade, was required for oncogenic progression. Collectively, this work shows that pathogenic fungi promote PDAC by driving the complement cascade through the activation of MBL.
It would seem that these findings provide the basis for yet another approach to early detection, namely analysis of gut mycobiomes, and perhaps even prevention of PDAC by dietary modulation of gut mycobiomes.