- PCR Amplification Was First Published on December 20, 1985
- There Are Now ~500,000 Journal Publications Indexed to PCR
- Next-Generation Sequencing Is Enabled by PCR

Introduction
The idea of choosing a day to celebrate the polymerase chain reaction (PCR) traces back to DNA Day, which is celebrated on April 25th each year. DNA Day commemorates the publication of the double helix structure of DNA by Watson & Crick in Nature in 1953. As chronicled in Wikipedia, DNA Day was first celebrated on April 25th, 2003 in the United States by a proclamation of Congress as a one-time event, but it soon became an international yearly phenomenon. Past blogs about DNA Day in the Zone can be accessed at this link.
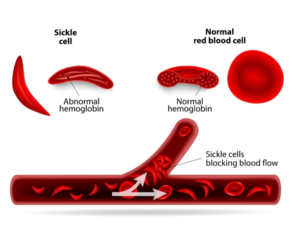 Appreciation for DNA as the molecular basis or “blueprint” of genetics evolved largely due to the co-evolution of various methods for the manipulation and analysis of DNA at the single-nucleotide level, for entire genomes. Among these DNA-based methods, PCR has arguably been the most enabling. First reported by Saiki et al. in Science on December 20th, 1985, PCR was discussed in an article titled Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia.
Appreciation for DNA as the molecular basis or “blueprint” of genetics evolved largely due to the co-evolution of various methods for the manipulation and analysis of DNA at the single-nucleotide level, for entire genomes. Among these DNA-based methods, PCR has arguably been the most enabling. First reported by Saiki et al. in Science on December 20th, 1985, PCR was discussed in an article titled Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia.
Since the Saiki et al. publication in 1985, which has been cited more than 11,300 times, PCR continues to be increasingly used for a wide variety of diverse applications, as indicated by this chart of ~500,000 publications indexed to the phrase “polymerase chain reaction” in PubMed. In addition, the same phrase is found in text searches of ~86,000 patents in the United States Patent and Trademark Office database, as well as in ~2,870,000 items in Google Scholar.
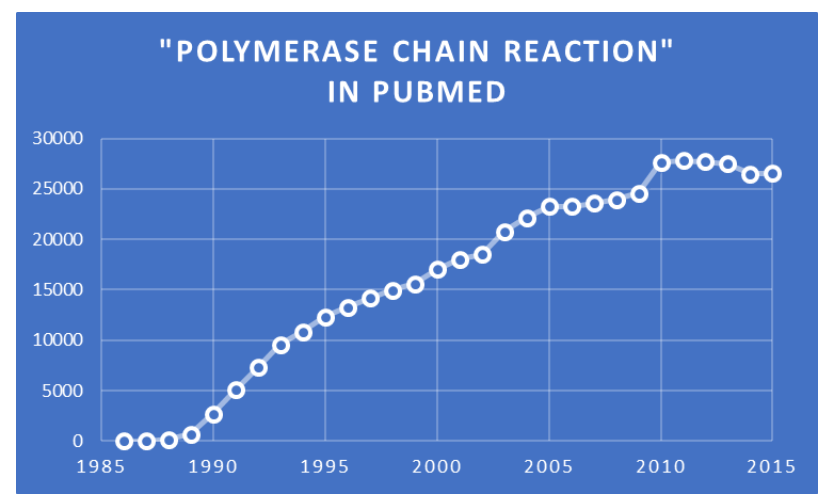 Chart of PubMed search results for the phrase “polymerase chain reaction” by Jerry Zon.
Chart of PubMed search results for the phrase “polymerase chain reaction” by Jerry Zon.
While the Saiki et al. publication date of December 20th will likely be officially recognized by some organization someday in the future, the Zone proposes that recognition is deserved now. This inaugural blog for PCR Day will briefly revisit the invention of PCR as a historical starting point, and then fast-forward, focusing on some examples of single-molecule amplification—the frontrunner in sensitivity—enabled by the 2n-amplification power of PCR.
The Inventor and Invention of PCR

Although the December 20th, 1985 report in Science by Sakai et al. was the first public disclosure of PCR, it tells nothing about the “who and how” of PCR discovery. These elements are best revealed by the inventor of PCR, Kary B. Mullis. His personal account titled The Unusual Origin of the Polymerase Chain Reaction, which was published in Scientific American in April 1990, is a “must read” for anyone who uses PCR. Some of the “unusual” aspects will be described below.
In 1983 at Cetus Corporation, Mullis was—in his words—“puttering around with oligonucleotides” in search of “a technique for easily determining the identity of the nucleotide at a given position in a DNA molecule.” The approach he was thinking about was essentially a variation of what nowadays is called polymerase-mediated single-base extension of an oligonucleotide primer using dideoxynucleotide triphosphate (ddNTP) terminators to identify single-nucleotide polymorphisms (SNPs) or mutations.
But the story gets even more interesting. Mullis says that “[f>
or the next few weeks I described the idea to anyone who would listen [but>
no one was particularly enthusiastic about it.” Undeterred, he did an initial experiment and got the expected gel result late that evening, when he bumped into Albert Halluin, the patent attorney for Cetus. Mullis told him about the idea of PCR and the results. Halluin “agreed that it was significant…[and>
was even a little excited and suggested that I get to work on the experiment and write a patent disclosure.”

Numerical Perspective for a Single Molecule

I am sure you get the point by now: working with only one molecule is exceedingly difficult. Common analytical methods for nucleic acids, such as gel electrophoresis and UV-visible spectroscopy, are generally applicable for umol to pmol quantities of analytes, and working with one molecule would not be possible for most scientists if it were not for PCR amplification.
Single-Molecule Amplification by PCR
The first report on single-molecule amplification by PCR was published in 1998 by Ohuchi et al., in a paper titled In vitro method for the generation of protein libraries using PCR amplification of a single DNA molecule and coupled transcription/translation. In this publication, the researchers coined the term ‘single-molecule PCR’ (smPCR). Single molecules of DNA template were obtained by serial dilution, and randomized three-base tags were incorporated into one of the primers for sequence-based evidence to address the question they posed: “Could a single molecule of DNA really be amplified?”
Data obtained by Ohuchi et al. indicated that the answer was yes; however, problems with non-specific amplification had to be addressed, and still are problematic for PCR despite its widespread usage. Unwanted primer dimers and mis-priming products, investigated early on at Cetus, are commonly formed under low stringency hybridization conditions that exist prior to PCR during sample preparation at ambient temperature, and during the initial heat-denaturing ramp of the PCR process. Once formed, these off-target products can compete with amplification of the desired DNA target and significantly decrease the efficiency of the method.
Over the years, various approaches have been investigated to mitigate these problems, as discussed by Koukhareva and Lebedev in a publication that describes the development of TriLink’s novel heat-reversible 3′-protected derivatives of dNTPs, depicted here, as substitutes for natural dNTPs in PCR. Since 3′-protected dNTPs are either non-substrates or terminating substrates for thermally stable DNA polymerases, such as Taq, they do not support primer

extension/elongation at low stringency conditions during PCR sample preparation, when PCR artifacts such as primer dimers and mis-priming products can form. After hot-start cycling, PCR proceeds with much improved specificity, especially in highly multiplexed PCR reactions. These modified dNTPs and various kits containing them are available from TriLink as CleanAmp® Hot Start PCR products.
Digital PCR
Digital PCR (dPCR) has revolutionized the way in which PCR is used for quantification. PCR carries out one reaction per single sample. dPCR also does this, but the single sample is separated into a large number of partitions, and the reaction is carried out in each partition individually, as depicted here. This separation allows for a more reliable collection and sensitive measurement of nucleic acid amounts.
Details on dPCR and its many applications can be found at this link. Among the benefits and applications of dPCR, absolute quantification really stands out. dPCR enables the absolute and reproducible quantification of target nucleic acids at single-molecule resolution. Unlike analogue quantitative PCR (qPCR), absolute quantification by dPCR does not require a standard curve.
dPCR also has a greater tolerance for inhibitor substances and PCR assays that amplify more inefficiently, when compared to qPCR. For example, dPCR can quantify specific sequences from contaminated genetically modified organisms (GMOs) in food, viral loads in the blood, biomarkers of neurodegenerative disease in cerebral spinal fluid, and even fecal contamination of drinking water.
Single-Molecule Amplification for Improved Bisulfite Sequencing
The treatment of DNA with bisulfite, which converts C to U but leaves 5-methyl-C unchanged, forms the basis of many analytical techniques for DNA methylation analysis. Many techniques exist for measuring the methylation state of a single CpG, but for analysis of an entire region, cloning and sequencing has been the gold standard. However, biases in PCR amplification and in cloning can skew the results.
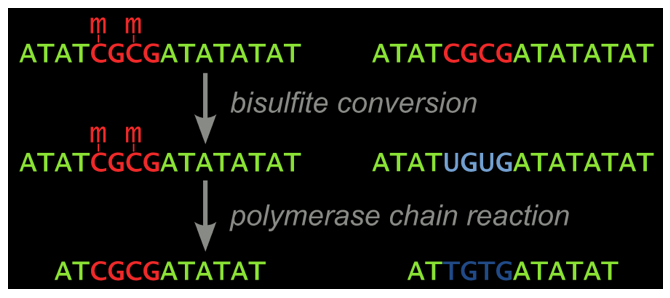
In a publication in Anal. Biochem., Chhibber and Schroeder hypothesized that smPCR would eliminate the amplification bias, as competition between templates that amplify at different efficiencies no longer exists. The amplified products can be sequenced directly, thus eliminating cloning bias. They demonstrated this accurate and unbiased approach by analyzing a sample that was expected to contain a 50:50 ratio of methylated to unmethylated molecules: a region of the X-linked FMR1 gene from a human female cell line. Sequencing smPCR products gave an expected methylated to unmethylated ratio of 48:52, whereas conventional cloning and sequencing gave a biased ratio of 72:28.
Single-Molecule Amplification for Next-Generation Sequencing
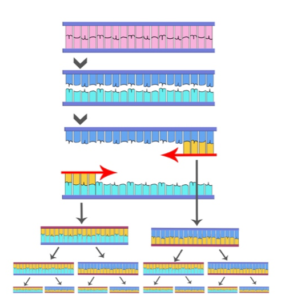
Next-Generation DNA Sequencing Methods by Elaine R. Mardis, a well-known sequencing expert, provides an excellent review of various highly multiplexed sequencing approaches that have come to be collectively known as next-generation sequencing (NGS). Several of these methods employ single-molecule PCR, either in emulsion or on a surface, as exemplified here for the widely used Illumina systems. According to Mardis, the single-molecule amplification step for the Illumina Genome Analyzer starts with an Illumina-specific adapter library (blue), and takes place on the oligo primer-derivatized surface of a flow cell, as depicted here (green and red primers).
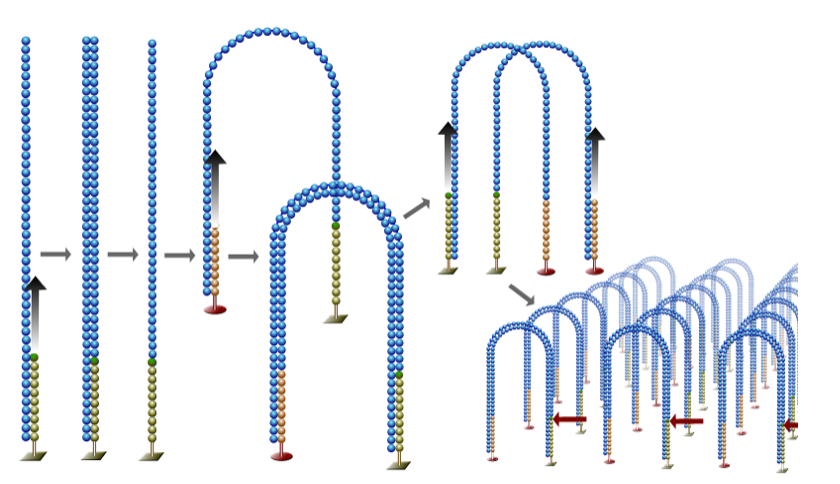
The overall process, called bridge amplification, is performed by an automated flow-cell device called a Cluster Station, as the PCR-like amplification leads to “clusters” of clonally pure DNA, depicted as tiny spots to be sequenced. Put another way, the DNA polymerase produces multiple DNA copies, or clusters, that each represent the single molecule that initiated the cluster amplification. A separate sample library can be added to each of the eight channels that comprise the device, and the same library can be used in all eight, or combinations thereof.
Each cluster contains approximately one million copies of the original fragment, which is sufficient for reporting incorporated bases at the required signal intensity for detection during sequencing, says Mardis. The Illumina system utilizes a sequencing-by-synthesis approach in which all four nucleotides are added simultaneously to the flow-cell channels, along with DNA polymerase, for incorporation into the oligo-primed cluster fragments.

Specifically, the color-coded nucleotides carry a base-unique fluorescent label, and the 3’-OH group is chemically blocked, making each incorporation is a unique event. An imaging step follows each base incorporation step, during which each flow cell lane is imaged by the instrument optics, as depicted here. After each imaging step, the 3’-blocking group is chemically removed to prepare each strand for the next incorporation by DNA polymerase. This series of steps continues for a specific number of cycles, as determined by user-defined instrument settings. In this manner, read lengths of 150—300 bases are achieved.
Concluding Comments

In the meantime, your comments are welcomed, as usual.