- Human DNA Misincorporates >1,000,000 Ribonucleotides Per Replication Cycle
- These Mistakes are Likely Biological Mysteries
- Four New Sequencing Methods May Demystify Why There’s “R in DNA”
When I came across a publication on the presence of RNA in DNA my initial reaction, frankly, was great surprise, if not outright disbelief. As the so-called “blueprint” of life, I reckoned that DNA is virtually sacred in terms of its chemical composition, albeit subject to base mutations as well as insertions and deletions of sequence. In other words, I had heretofore been under the impression that DNA’s repeating units are 100% deoxyribonucleotide (and conversely that RNA’s are ribonucleotides), thus giving DNA (and RNA) the eponymous name is has. So, I thought to myself, if that’s reportedly not the case for DNA, what are the facts and implications, i.e., is RNA in DNA just a rare “mistake” or is this yet another example of a “mystery” of Nature. Below is what I’ve learned about this revelation.
Fidelity: Complementarity vs. Discrimination
Faithful replication of the genome is essential for the propagation of cells, and multiple mechanisms have evolved to ensure the accurate copying of DNA during cell division. In general, replication fidelity refers to the preservation of base identity between genomic template (parent) and copied (daughter) strands of DNA. Replication enzymes (DNA polymerases) insert deoxyribonucleotides (dNTPs) at the 3′ end of the nascent strand that are complementary to the opposite base on the template strand: namely, complementarity of A-T and G-C base pairs. Proofreading activity of DNA polymerases as well as other, post-replicative enzymes act to recognize and repair mispaired dNMPs that are inserted during replication. In this way, there is multi-tiered protection against mutations, and the error rate of replication is kept extremely low.
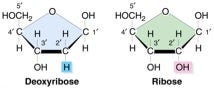
Apart from ensuring complementarity, another facet of replication fidelity is the discrimination of the sugar backbone of the nucleic acid strand (ribose rNTPs versus deoxyribose dNTPs) so that the correct sugar NTP is chosen. As seen from the structure above, these two classes have very similar structures, differing by the added presence of only a single oxygen in ribo vs. deoxy forms. Hence, polymerases are challenged with distinguishing between the two types of nucleotides, otherwise they risk inserting the incorrect substrate into the newly replicated nucleic acid strand. Since RNA moieties in what should be 100% DNA would seemingly lead to various problems, I was surprised to learn that ribonucleotide misincorporation represents the most common type of replication error and occurs quite frequently in normal cells. Even more surprising, as you will now read, there are some positive consequences to having “some Rs among the Ds,” so to speak.
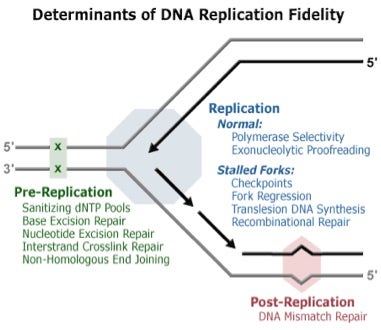 Mechanisms involved in DNA replication: pre-replication, replication and post-replication. Taken from DNA Replication Fidelity Group via Google Images.
Mechanisms involved in DNA replication: pre-replication, replication and post-replication. Taken from DNA Replication Fidelity Group via Google Images.
Frequency and Consequences of RNA in DNA
According to a recent review by Potenski & Klein, rates of ribonucleotide misincorporation are influenced by many factors: e.g. replicative polymerase, the rNTP in question, the surrounding sequence context and the ratios of rNTP to dNTP pools (which change, but typically involve ∼100-fold more rNTP than dNTP).
As such, sugar discrimination is a very important trait for DNA polymerases in this environment enriched for rNTPs over dNTPs. Studies of human Pol δ by Kunkel and coworkers found that in reactions containing nucleotide concentrations estimated to be present in mammalian cells, there was one rNTP incorporated per ∼2000 dNTPs—a result which predicts that human Pol δ may introduce >1,000,000 rNMPs into the genome per replication cycle.
Earlier results in analogous studies by Kunkel and coworkers using yeast led these investigators to state that “[t> he idea that rNMP incorporation into DNA may be more common than previously appreciated leads one to wonder about possible benefits of rNMP incorporation.”
- Mating type switching in haploid cells of the fission yeast Schizosaccharomyces pombe, which provides an example of ribonucleotide-based epigenetic imprint to differentiate sister chromatid strands.
- Utilization of ribonucleotides during non-homologous end joining (NHEJ), which repairs double-strand breaks in DNA, via 3’-ribonucleotide stimulation of DNA ligase IV.
- Signaling for DNA mismatch repair (MMR) via rNTP incorporation during replication to mark the nascent strands for MMR.
Global Mapping of Genomic Ribonucleotides
Having learned all of the above information that was completely new to me, the obvious next thoughts I had were questions of where in DNA were these RNA moieties, and were these locations random or grouped in ways suggesting function? Remarkably, and to this point, four completely independent studies of methods for mapping genomic ribonucleotides were contemporaneously published on January 26, 2015:
- Reijns et al. Lagging-strand replication shapes the mutational landscape of the genome (Nature). This method called emRiboSeq starts with fragmenting gDNA and then ligating adapters to each end. Ribonuclease H2 is then used to cut strands containing a ribonucleotide, and a second adapter is attached to the 3’ end of those ribonucleotide-containing strands. The second adapters are captured and their attached sequences are mapped to the genome.
- Clausen et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation (Nature Structural & Molecular Biology). This work by Kunkel’s group involves a similar approach called HydEn-seq, where they used a different enzyme to cleave ribonucleotide-containing DNA fragments, and captured the resultant pieces downstream rather than upstream.
- Koh et al. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA (Nature Methods). This method also cleaves ribonucleotide-containing DNA cleaves but relies on a different capture method that affords single-stranded DNA circles, which are then amplified and sequenced.
- Daigaku et al. A global profile of replicative polymerase usage (Nature Structural & Molecular Biology). This approach called Pu-seq involves treating ribonucleotide-containing DNA with alkali, which cleaves DNA at each of the ribonucleotides. The resultant fragments are then ligated to adaptors, amplified, and sequenced.
Although none of the methods have been compared head-to-head yet, all of the datasets are publicly available for interested researchers. Needless to say, these datasets are “dreams come true” for bioinformaticians, who will have many fun challenges writing algorithms to parse the data in attempts to conjure up possible functional rationale. Stay tuned on more insights into the “mystery” of Nature being revealed by yet another new application for deep sequencing, which I’ll call “ribonomics” or, more mundanely, “what’s the R in DNA for?”
As usual, your comments are welcomes.