Targeted Genome Engineering with Zinc-finger Nucleases, TALENs and CRISPR
Targeted genome editing tools such as meganucleases, zinc-finger nucleases, TALENs and CRISPR are among the hottest topics in cell and gene therapy. Dr. Anton McCaffrey, Principal Scientist at TriLink and expert in these areas, gives herein his overview after attending the recent American Society for Gene and Cell Therapy Meeting (May 2013) and the International Society for Stem Cell Research Meeting (June 2013) where there were a number of exciting talks discussing applications of this technology.

Dr. McCaffrey received his PhD in Biochemistry from the University of Colorado at Boulder in 1999. During his postdoctoral fellowship at Stanford he developed gene therapeutics for hepatitis B and C. He was then Assistant Professor at
University of Iowa where he focused on the role of microRNAs during the pathogenesis of hepatitis C virus and developed RNA interference and zinc-finger nuclease based therapeutics for treatment of hepatitis B virus. Now he manages the RNA Transcription product line at TriLink.
So what is targeted genome engineering with nucleases and why would you want to do this? The primary goal in cells or animals is to create a specific, localized double-stranded DNA break and then to: 1. correct the sequence of a defective targeted gene, 2. knock in a specific gene mutation to create a disease model or 3. knock out a gene. The basic idea is to rationally design artificial restriction enzymes that recognize a specific location within the DNA genome of a cell or organism and catalyze a double stranded break at this location (Figure 1).
In the first two cases, where the target gene is to be specifically edited, the nuclease(s) are co-transfected with an exogenous donor DNA molecule. This donor DNA contains arms, which share homology with the target loci and will direct homologous recombination at the targeted cut site. The sequence of the donor DNA replaces that of the endogenous locus at one or both alleles. So, for example, a wild-type donor sequence can be used to replace a mutated gene sequence to correct a genetic disease.
If one wishes to inactivate a gene using these technologies, an exogenous donor template is not included. In the absence of a donor, the cell uses non-homologous end joining to repair the double stranded break. At a high frequency, this process introduces deletions and insertions in the gene, which changes the reading frame and inactivates the gene.
Until the advent of these technologies, it was impossible to make transgenic animals other than mice. Using targeting genome engineering is now possible to make transgenic rats, pigs, ferrets and plants. As will be discussed below, advances in messenger RNA (mRNA)-based gene therapy are converging with advances in targeted genome engineering to enable efficient, yet transient expression of designer nucleases without risk of undesired integration of the nuclease expression vector.
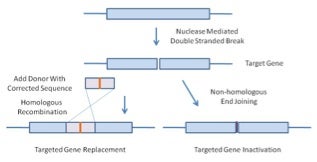
Figure 1. Nuclease Mediated Double Stranded Breaks Stimulate Homologous Gene Replacement or Targeted Gene Inactivation. If targeted nucleases are co-transfected with a homologous donor DNA fragment, homologous recombination replaces defective DNA with a corrected sequence (left). In the absence of a DNA donor fragment, non-homologous end joining repairs the break, but with frequent insertions and deletions, thus inactivating the gene.
Techniques for making targeted nucleases are rapidly evolving. Initial attempts to engineer designer restriction nucleases to target new sequences revolved around changing the specificity of naturally occurring nucleases such as meganucleases. Meganucleases are restriction enzymes with long recognition sites (12-40 nucleotides). These nucleases could be engineered to recognize related sequences in genomes and cleave them. However, only a small number of sites could be targeted using this approach (refs A-C).
Zinc-finger nucleases (ZFNs) were the next major advance in the field. Zinc fingers are the most common DNA binding motif in mammalian transcription factors. These sequence specific binding domains can be engineered to bind to novel DNA sequences. Zinc-fingers can be turned into nucleases by fusing them to non-specific cleavage domains, such as the FokI nuclease. FokI cleaves as a dimer, so pairs of ZFNs are designed to bind to adjacent sites in the genome to allow FokI dimer formation and double stranded DNA cleavage (Figure 2). A number of laboratories published design rules that serve as a starting point to engineer ZFNs with novel DNA binding specificities (refs N-R). In reality, actual binding specificity is context dependent. Several selection protocols in cells also exist for identifying novel ZFNs. ZFNs have been successfully used to modify the genomes of Drosophila, C. elegans, zebrafish and rats (refs D-M). However, identification of functional ZFNs remains challenging and most ZFNs have emerged from a small number of laboratories with specialist skills.
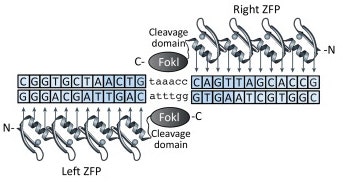
Figure 2. Zinc-Finger Nucleases Bind as Dimers to Cut Double Stranded DNA. Adapted from Gaj et al.Trends Biotechnol. 2013 May 8. [Epub ahead of print>
In the last few years, TALENs have emerged as a more generally accessible alternative to ZFNs. Like ZFNs, TALENs utilize a modular DNA binding motif (TALE) that can be modified to introduce new DNA binding specificities. TALENs consist of multiple repeat variable diresidues (RVDs) which each specify binding to a single nucleotide (Refs S-U). TALEN arrays are made by stringing together RVDs in a specific order to provide specificity and binding affinity to novel DNA sequences. Commonly, engineered TALE sequences are fused to non-specific cleavage domains such as FokI. As with ZFNs, TALENs function as pairs bound to adjacent DNA sequences. Unlike ZFNs, TALENs are not as prone to sequence context effects, which greatly complicate the de novo design of ZFNs. This has made them much more accessible to the general scientific community. A number of groups have published TALEN assembly protocols that allow assembly of these repetitive sequences, including one popular open source assembly method is known as Golden Gate (Refs V-Y).

Figure 3. TALENs Bind as Dimers to Cut Double Stranded DNA. Adapted from Gaj et al.Trends Biotechnol. 2013 May 8. [Epub ahead of print>
The newest kid on the genome-engineering block is CRISPR. CRISPR is a bacterial immune system in which bacteria sample the DNA of pathogens, integrate foreign DNA into their genome in specialized repeat structures, and then use these sequences to produce Guide RNAs that direct cutting of homologous pathogenic DNA sequences. To some degree this is reminiscent of RNA interference in mammals. Once the target site has been delineated by the RNA guide sequence, Cas proteins (CRISPR-associated proteins) do the cutting. A number of groups have adapted this system to create RNA directed genome engineering tools (refs Z-CC) (Figure 4). This new system has generated considerable interest since recognition of the target DNA sequence to be cut is RNA mediated rather than protein mediated. DNA cleavage is carried out by the expressed Cas9 protein. With ZFNs and TALENs, if you wish to target a new site, you have to identify and synthesize two new proteins. With CRISPR, you use the same Cas9 protein each time and just alter the sequence of the guide RNA. Stay tuned for head to head comparisons of the efficiency and specificity of ZFNs, TALENs and CRISPR that are about to be published.
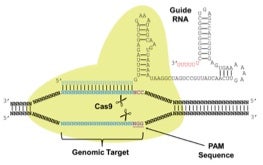
Figure 4. CRISPR is an RNA Guided Genome Engineering System. Figure adapted from DiCarlo et al. Nucleic Acids Research, 2013, Vol. 41, No. 7.
Modified mRNA for Transient Expression in Genome Engineering
In each of the three systems described above, one needs to express one or two proteins inside cells or an organism. Plasmids and viral vectors have been used to achieve this, but these carry a risk. Double stranded DNA breaks catalyze insertion of DNA at the cut site. At some substantial frequency, the protein expression vectors can integrate at the cut site. These vectors necessarily carry eukaryotic promoters, which can lead to continuous expression of the nuclease or the expression of previously silent sequences. For clinical applications this can be a major issue. One also needs to consider off-target cleavage of by engineered nucleases. Since ZFNs have been around longer than TALENs or CRISPR, the most data exists for ZFNs. It is clear that ZFNs can cut at pseudo-sites that resemble the chosen target site.For this reason, transient expression of nucleases is desirable. Many in the ZFN and TALEN field have moved to expression of these nucleases from synthetic mRNAs because they are transient and have no risk of insertion. Synthetic mRNAs, which mimic fully processed, capped and polyadenylated mRNAs, can be produced in large quantities by in vitro transcription. Transfected mRNAs made with adenine, cytosine, guanine and uracil are recognized as pathogens by innate immune sensors such as Toll-like receptors, RIG-I and PKR. Kariko et al. showed that mRNAs could be made much less immunogenic and non-toxic by substitution of cytosine and uridine with 5-methylcytosine and pseudouridine (ref DD). Custom syntheses of milligram to gram amounts of 5-methylcytosine and pseudouridine modified mRNAs can be ordered from TriLink BioTechnologies. Cas9 mRNA is also available as a catalog item.
Conclusions
In recent years, designer genome engineering has gone from dream to reality. New editing systems are taking this from the realm of a few elite laboratories and companies to democratizing it for the masses. Concurrent advances in mRNA gene therapy are providing safe and effective delivery systems for expressing the necessary components in cells and animals. There is now huge interest in using targeted genome engineering in patient derived somatic cells and stem cells. Rather than simply knocking genes in or knocking them out, we may now be able to actually correct monogenic genetic disorders. Clinical trials are currently under way to determine if ZFNs can be used to inactivate the CCR5 HIV co-receptor to make patient T-cells immune to HIV. These technologies will also enable facile creation of disease models in species other than mice. The future is bright for targeted genome engineering. That’s the buzz on the cut.
A sincere thanks to Anton McCaffrey for providing this update on truly exciting trends in nucleic acid-based technologie. As always, I welcome comments and discussions.
References
A. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Cohen-Tannoudji M, Robine S, Choulika A, et al. Mol Cell Biol 1998;18:1444-8.
B. The yeast I-Sce I meganuclease induces site-directed chromosomal recombination in mammalian cells. Choulika A, Perrin A, Dujon B, Nicolas JF. C R Acad Sci III 1994;317:1013-9.
C. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Choulika A, Perrin A, Dujon B, Nicolas JF. Mol Cell Biol 1995;15:1968-73.
ZFNs modifying different organisms
D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Genetics 2006;172:2391-403.
E. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Beumer KJ, Trautman JK, Bozas A, et al. Proc Natl Acad Sci U S A 2008;105:19821-6.
F. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Bibikova M, Golic M, Golic KG, Carroll D. Genetics 2002;161:1169-75.
G. Genetic Analysis of Zinc-finger Nuclease-induced Gene Targeting in Drosophila. Bozas A, Beumer KJ, Trautman JK, Carroll D. Genetics 2009.
H. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Morton J, Davis MW, Jorgensen EM, Carroll D. Proc Natl Acad Sci U S A 2006;103:16370-5.
I. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Doyon Y, McCammon JM, Miller JC, et al. Nat Biotechnol 2008;26:702-8.
J. Zinc finger-based knockout punches for zebrafish genes. Ekker SC. Zebrafish 2008;5:121-3.
K. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN). Foley JE, Yeh JR, Maeder ML, et al. PLoS ONE 2009;4:e4348.
L. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Nat Biotechnol 2008;26:695-701.
M. Knockout rats via embryo microinjection of zinc-finger nucleases. Geurts AM, Cost GJ, Freyvert Y, et al. Science 2009;325:433.
ZFN design rules
N. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Segal DJ, Dreier B, Beerli RR, Barbas CF, 3rd. Proc Natl Acad Sci U S A 1999;96:2758-63.
O. Insights into the molecular recognition of the 5'-GNN-3' family of DNA sequences by zinc finger domains. Dreier B, Segal DJ, Barbas CF, 3rd. J Mol Biol 2000;303:489-502.
P. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. Liu Q, Xia Z, Zhong X, Case CC. J Biol Chem 2002;277:3850-6.
Q. Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors. Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF, 3rd. J Biol Chem 2001;276:29466-78.
R. Development of zinc finger domains for recognition of the 5'-CNN-3' family DNA sequences and their use in the construction of artificial transcription factors. Dreier B, Fuller RP, Segal DJ, et al. J Biol Chem 2005;280:35588-97.
TALENS
Breaking the code of DNA binding specificity of TAL-type III effectors. S. Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., Lahaye, T., Nickstadt, A. and Bonas, U. Science 2009;326,1509-12.
T. The crystal structure of TAL effector PthXo1 bound to its DNA target. Mak, A.N., Bradley, P., Cernadas, R.A., Bogdanove, A.J. and Stoddard, B.L. Science 2012;335, 716-9.
U. A simple cipher governs DNA recognition by TAL effectors. Moscou, M.J. and Bogdanove, A.J. Science 2009;326,1501.
Golden gate
V. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Cermak, T., Doyle, E.L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., Baller, J.A., Somia, N.V., Bogdanove, A.J., Voytas, D.F., Geissler et al. Nucleic Acids Res 2011;39,e82.
W. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Li, T., Huang, S., Zhao, X., Wright, D.A., Carpenter, S., Spalding, M.H., Weeks, D.P. and Yang, B., Morbitzer et al. Nucleic Acids Res 2011;39, 6315-25.
X. A modular cloning system for standardized assembly of multigene constructs. Weber, E., Engler, C., Gruetzner, R., Werner, S. and Marillonnet, S. PLoS One 2011;6, e16765.
Y. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Zhang, F., Cong, L., Lodato, S., Kosuri, S., Church, G.M. and Arlotta, P. Nat Biotechnol 2011;29,149-53.
CRISPR
Z. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Jinek, M; Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science 2012;PMID 22745249.
AA. Multiplex genome engineering using CRISPR/Cas systems. Cong, Le; Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Science 2013;PMID 23287718.
BB. RNA-guided human genome engineering via Cas9. Mali, P; Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Science 2013;PMID 23287722.
CC. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Wang, H; Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. Cell 2013;PMID 23643243.
Modified mRNA
DD. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Mol Ther. 2008;Nov;16(11):1833-40.






