
X-shredding: Man-Made Male
March 2018
In the inaugural edition of The CRISPR Journal, researchers from the University of Perugia published a method called X-Shredding, which identifies key loci of the X-chromosome in order to bias sex ratio.
X-shredding is a technique that utilizes the CRISPR-Cas9 system to target the X-chromosome. This leads to endonuclease-mediated X-chromosome shredding during spermatogenesis, which manipulates the sex ratio towards males.
There has been significant interest in this method because it may provide a feasible alternative to pesticides in the control of insect populations. Blood-feeding insects are responsible for more than half a million deaths each year worldwide. The Food and Agriculture Organization of the United Nations reports that herbivorous insects are responsible for the destruction of one fifth of the world’s crops annually.
"However, to build synthetic X-shredders based on CRISPR, the selection of gRNA targets in the form of high-copy sequence repeats on the X chromosome of a given species is difficult, since such repeats are not accurately resolved in genome assemblies and cannot be assigned to chromosomes with confidence," the researchers note.
To address this hurdle, the researchers built a bioinformatics pipeline, designed to require only raw whole genome sequencing data with nominal filtering and error correction.
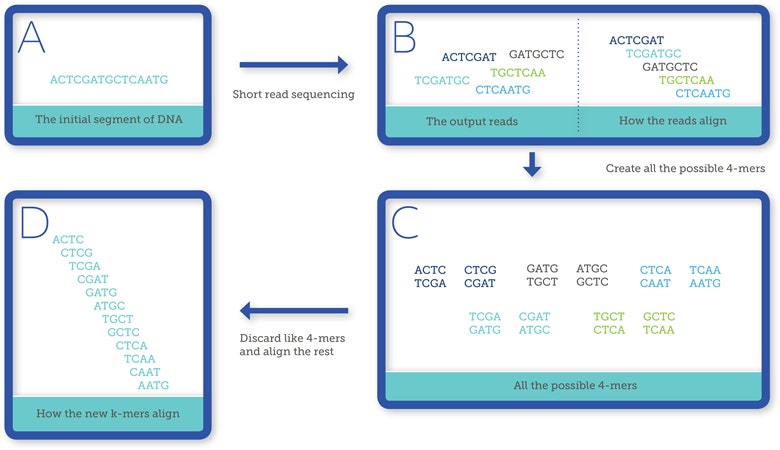
This process enables the identification of highly abundant X-specific sequences in the absence of genome assembly. The method is termed redkmer, because it works through the repeat extraction and detection of target sequences based on k-mers. In bioinformatics, k-mer refers to all the possible subsequences (of length k) from a read obtained through DNA sequencing (Figure 1).
To summarize, the redkmer process uses a combination of long-read and single-molecule sequencing, combined with short-read WGS data from females and males, to identify sequences specific to the X-chromosome (See Figure 2 for schematic of full process).

The identified sequences are introduced into a DNA cassette with sequences for Cas9 and flanking sequences. This allows for integration into the genome of the target species.
The research team showed evidence that the redkmer method results in efficient and reliable identification of X-chromosome specific sequences that can manipulate the sex ratio in mosquitos (Anopheles gambiae) through X-shredding.
Although this approach appears feasible, there is already concern as to whether the spread of the modified DNA cassette can really be controlled once the genetically modified insects are released into the wild.
In a recent article published in EMBO, the authors called for policy makers to ensure that proper regulations are established and potential impacts on the environment are adequately assessed.
Relevant products: TriLink offers custom synthesis of gRNA oligos as well as several standard and custom variations of Cas9 mRNA. Please see our comprehensive list of stocked constructs here. If you need a unique Cas9 analog or incorporation of specific base modifications, please contact us to discuss our custom synthesis service.
Have a question? Visit Ask An Expert.
© 2018 TriLink BioTechnologies. | All Rights Reserved

