Through co-delivery of mRNA-encoded CRISPR-associated nuclease and single guide RNA (sgRNA) specific for a target locus in the genome, lipid nanoparticles (LNPs) have successfully achieved in vivo gene editing in both preclinical and clinical studies (Gillmore et al.). However, compared to mRNA vaccines, mRNA-encoded CRISPR therapeutics require up to 1,000-fold-higher mRNA dosage (or protein expression) to reach a therapeutic threshold (Rohner et al.), which raises higher requirements for the potency and safety of LNPs. Therefore, there is much interest in developing next-generation ionizable lipids (ILs) – a critical component of LNPs that governs the efficacy and biocompatibility. Moreover, new methods for simple, robust, and scalable synthesis of these ILs are valuable for practical applications.
This blog discusses the development of a plug-and-play assembly strategy for the combinatorial synthesis of biodegradable ILs recently published by researchers (Han et al.) at the University of Pennsylvania. Compared to benchmark ILs, the lead IL — synthesized in one step from an amine and a dialkyl maleate — demonstrates superior performance in delivering various CRISPR mRNA therapeutics in wild-type and transgenic mice.
The study used TriLink firefly luciferase (FLuc) mRNA (5moU)as a reporter for screening ILs and TriLink Cas9 mRNA (5moU) for gene knockout. mRNAs encoding either an adenine base editor or a cytidine base editor were co-transcriptionally prepared with TriLink CleanCap® and N1-methylpseudouridine-5’-triphosphate reagents. The following sections briefly summarize each of these TriLink product applications.
Plug-and-play assembly of ILs and in vitroscreening
Han et al. devised a two-component combinatorial chemistry strategy for “plug-and-play” assembly of biodegradable ILs using amines/thiols and dialkyl maleates (Figure 1).
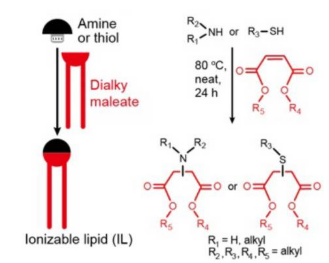
FIGURE 1.Plug-and-play assembly of biodegradable ILs by Michael addition between amines/thiols (black) and dialkyl maleates (red). Image taken from Han et al. and used under CC-BY-NC-ND 4.0 license.
This Michael addition reaction has notable advantages: easily accessible building blocks, a simple experimental procedure, avoidance of solvent or catalyst, and a high product yield. ILs with asymmetric tails can be easily prepared from asymmetric dialkyl maleates, which expands the structural diversity of ILs.
500 ILs were combinatorially synthesized using 50 amines/thiols and 10 dialkyl maleates. These ILs were individually formulated into FLuc mRNA-LNPs, with other conventional lipid excipients, and then subjected to high-throughput in vitro screening. HepG2 cells (a human hepatocellular carcinoma cell line) were treated with each test sample at a low mRNA dose (3 ng/well) to avoid cytotoxicity, and FLuc expression was measured after 24 hours.
The publication by Han et al. discusses details for several trends among IL structures vs. relative levels of luciferase activity. However, for the purpose of this blog, it needs only be mentioned that, of the initial 500 candidate ILs, 200 amine-derived ILs affording relatively high levels of luciferase fluorescence were selected for the in vivo screening outlined next.
Comparative IL screening in vivo
To accelerate the identification of optimal amines and reduce the usage of animals, Han et al. used a batch screening method. Briefly, FLuc mRNA-LNPs with the same amine structure were pooled into 20 batches (10 LNPs per batch), which were intravenously (i.v.) injected into mice at a FLuc mRNA dose of 0.1 mg/kg. Four hours later, mice were intraperitoneally injected with D-luciferinsubstrate for bioluminescence imaging using an in vivo imaging system.
Whole-body total bioluminescence was quantified and normalized to mice similarly treated with C12–200 LNPs, a potent benchmark for the delivery of mRNA therapeutics and gene editors (Love et al.). These in vivo batch screening results showed that two amines, designated 8 and 5, were the best performers and achieved higher (3.6-fold) and slightly lower (0.6-fold) transfection efficiency relative to C12–200 LNP, respectively.
Individual screening in mice was then performed with each of nine amine 8-derived ILs (Figure 2). To ensure an unbiased comparison, these ILs were each purified and formulated into FLuc mRNA-loaded LNPs using a microfluidic device. In vivo results demonstrated that LNPs formulated with amine 8 having short multiple tails achieved comparable or higher transfection efficiency than the benchmark C12–200 LNP, while those with increased tail and branch length showed decreased transfection activity. These results were consistent with those of a previous study (Hajj et al.) wherein potent ILs often contain multiple short tails.
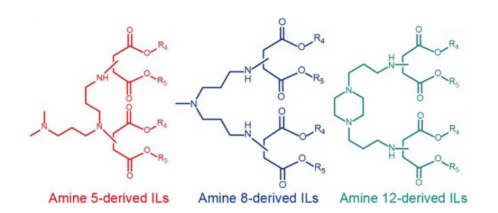
FIGURE 2. Structures of amine 5-, 8-, and 12-derived ILs. Image taken from Han et al. and used under CC-BY-NC-ND 4.0 license.
Similar screening of five individual amine 5-derived ILs (Figure2) with multiple short tails also exhibited comparable or superior activity relative to the benchmark C12–200 LNP. On the other hand, in vivo testing of five amine 12-derived ILs (Figure 2) with multiple short tails gave results that were slightly inferior to the C12–200 LNP benchmark.
Utility of ILs to deliver CRISPR mRNA for in vivo gene editing
Inspired by the ease of synthesis and highly efficient FLuc mRNA delivery in vivo, Han et al. tested the utility of ILs to deliver CRISPR mRNA therapeutics for in vivo gene editing. Five superior ILs from amine 5 and five superior ILs from amine 8 were each screened for liver genome editing by co-delivering Cas9 mRNA and sgRNA targeting the transthyretin (TTR) gene in mice. Five inferior amine 5-, 8-, or 12-derived ILs from the above-mentioned FLuc mRNA screen were also tested.
Encouragingly, all ten superior ILs achieved more than 27% on-target editing efficiency, measured by deep sequencing of TTR amplicons and determination of the on-target indel frequency. The lead-formulated LNP termed 5D8 (Figure 3) achieved the highest on-target editing efficiency (approximately 61%), which out-performed C12–200 LNP (51%) and other industry benchmark LNPs, namely, SM-102 LNP (20%), DLin-MC3 LNP (approximately 20%), DLin-MC3 LNP (approx. 5%), and LP-01 LNP (approx. 3%). Correspondingly, serum TTR protein was reduced by approx. 90% in mice treated with 5D8 LNP. In contrast, all inferior ILs barely facilitated liver gene editing and serum TTR reduction.

FIGURE 3. Top-performing 5DB IL. Image taken from Han et al. and used under CC-BY-NC-ND 4.0 license.
Of note, 5D8 LNP was also shown to be highly potent for delivery of short-interfering RNA, achieving approximately 100% reduction of serum TTR after delivery of TTR siRNA at a very low dose (0.05 mg/kg), which is superior to that previously reported for industry benchmark DLin-MC3 LNP under the same experimental conditions. Han et al. suggested that one possible reason for the high potency of 5D8 LNP could be its enrichment of apolipoprotein E on the surface, which is implicated in the active targeting of hepatocytes (Akinc et al.)
The hepatotoxicity of 5D8 LNP following in vivo gene editing was next investigated. Importantly, there was no observable increase in the levels of alanine transaminase (ALT) and aspartate aminotransferase (AST) live-toxicity markers following 5D8 LNP treatment. In contrast, treatment with the benchmark C12–200 LNP resulted in a slight elevation of both ALT and AST levels. Taken together, it was concluded that 5D8 is a superior ionizable lipid that enables potent mRNA delivery and gene editing with a favorable safety profile.
Utility of ILs for in vivo adenine and cytidine base editing
Han et al. next investigated whether the top-performing 5D8 LNP could serve as a universal delivery platform for other mRNA-based gene editing tools, such as CRISPR base editors. Unlike CRISPR/Cas9-based gene editors, base editors enable precise and efficient base changes without the introduction of double-stranded DNA breaks. Base editing can be used to permanently turn off disease-associated genes or correct pathogenic point mutations to treat single-nucleotide genetic diseases (Porto et al.).
The ability of 5D8 LNP to deliver adenine base editors (ABEs), which convert A•T base pairs to G•C base pairs, was studied first. Mice were i.v. injected with LNPs encapsulating mRNA encoding an ABE version 8.8m (ABE8.8) and a sgRNA targeting the PCSK9 (proprotein convertase subtilisin/kexin type 9) gene, a well-validated therapeutic target for the treatment of atherosclerotic cardiovascular disease (Lee et al.).Deep sequencing of PCSK9 amplicons showed that 5D8 LNP induced approximately 42% liver PCSK9 base-editing efficiency with a concomitant approximately 74% reduction of PCSK9 serum protein. In contrast, C12–200 LNP resulted in significantly lower PCSK9 editing efficiency (approximately 23%) and less serum PCSK9 reduction (approximately 52%) at the same dose.
Next explored was the potential of 5D8 LNP for gene editing therapy in mice with a genetic disease. Hereditary tyrosinaemia type 1 (HT1) results from a loss of function mutation in the FAH (fumarylacetoacetate hydrolase) gene, blocking the tyrosine catabolic pathway. Pharmacological inhibition of the upstream HPD (4-hydroxyphenylpyruvic acid dioxygenase) enzyme with nitisinone or knockout of HPD gene by base editing prevents the build-up of toxic metabolites and lethal liver failure.
LNPs comprising mRNA encoding a cytosine base editor, CBE version 4max (CBE4max), and a sgRNA targeting the HPD gene to introduce a C → T nonsense mutation were co-formulated and i.v. administered into adult FAH–/– mice. Deep sequencing of HPD amplicons showed that 5D8 LNP resulted in approx. 6.5% on-target editing, which was significantly higher than that (approx. 1.6%) achieved by C12–200 LNP. Consequently, mice receiving 5D8 LNP had a greater median survival time after nitisinone withdrawal (32 days), in comparison with PBS-treated group (5 days) and C12–200 LNP-treated group (18 days). Taken together, these results indicate that 5D8 LNP is a universal platform for efficient gene editing applications.
Concluding Comments
In summary, Han et al. developed a robust and straightforward combinatorial chemistry for the plug-and-play assembly of biodegradable ILs. Amine 5- and 8-derived ILs with multiple short tails showed superior in vivo mRNA delivery capability, among which 5D8 was identified as the lead candidate for liver gene editing with good tolerability. At clinically relevant doses, 5D8 LNP achieved higher gene editing efficiency or base editing efficiency than benchmark LNPs in wild-type and transgenic mice.
Your comments are welcome, as usual.







